- Current
- Browse
- Collections
-
For contributors
- For Authors
- Instructions to authors
- Article processing charge
- e-submission
- For Reviewers
- Instructions for reviewers
- How to become a reviewer
- Best reviewers
- For Readers
- Readership
- Subscription
- Permission guidelines
- About
- Editorial policy
Articles
- Page Path
- HOME > Diabetes Metab J > Ahead-of print > Article
-
ReviewBasic Research Roles of Histone Deacetylase 4 in the Inflammatory and Metabolic Processes
-
Hyunju Kang1
 , Young-Ki Park2, Ji-Young Lee2, Minkyung Bae3
, Young-Ki Park2, Ji-Young Lee2, Minkyung Bae3
-
DOI: https://doi.org/10.4093/dmj.2023.0174
Published online: March 22, 2024
- 627 Views
- 33 Download
1Department of Food and Nutrition, Keimyung University, Daegu, Korea
2Department of Nutritional Sciences, University of Connecticut, Storrs, CT, USA
3Department of Food and Nutrition, Yonsei University, Seoul, Korea
- Corresponding author: Minkyung Bae Department of Food and Nutrition, Yonsei University, 50 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea E-mail: minkyung.bae@yonsei.ac.kr
• Received: June 5, 2023 • Accepted: February 7, 2024
Copyright © 2024 Korean Diabetes Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
- Histone deacetylase 4 (HDAC4), a class IIa HDAC, has gained attention as a potential therapeutic target in treating inflammatory and metabolic processes based on its essential role in various biological pathways by deacetylating non-histone proteins, including transcription factors. The activity of HDAC4 is regulated at the transcriptional, post-transcriptional, and post-translational levels. The functions of HDAC4 are tissue-dependent in response to endogenous and exogenous factors and their substrates. In particular, the association of HDAC4 with non-histone targets, including transcription factors, such as myocyte enhancer factor 2, hypoxia-inducible factor, signal transducer and activator of transcription 1, and forkhead box proteins, play a crucial role in regulating inflammatory and metabolic processes. This review summarizes the regulatory modes of HDAC4 activity and its functions in inflammation, insulin signaling and glucose metabolism, and cardiac muscle development.
Highlights
- Alterations in chromatin structure impact the induction or repression of gene transcription by regulating the accessibility of transcription factors or other DNA-binding proteins to DNA [1]. Histones are major chromatin components and assemble with DNA to form nucleosomes, the smallest unit of chromatin [2]. Histone modifications, including phosphorylation, carbonylation, and acetylation, alter chromatin structure, altering replication, transcription, and DNA damage repair [3]. Histone acetylation by histone acetyltransferases leads to nucleosomal relaxation, favoring transcription factors to bind to DNA for gene induction. In contrast, histone deacetylation by histone deacetylases (HDACs) condenses chromatin structure for transcriptional repression. The reversible acetylation/deacetylation in lysine residues of histones can regulate biological pathways by activating or repressing transcription [1,4,5]. In addition to histones, there are several non-histone proteins that may undergo reversible deacetylation by HDACs [6]. For instance, HDACs deacetylates specific transcription factors involved in a wide range of biological processes, including apoptosis, autophagy, DNA repair, cell differentiation, and proliferation [6,7].
- HDACs are categorized into four classes according to their sequence homology and their dependence on cofactors (Fig. 1) [8]. The class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) possess homology to yeast Rpd3 protein [9]. The class II HDACs possess homology to yeast I1 protein [9], and they are further divided into two subclasses, class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10). The class III HDACs (sirtuin 1 [SIRT1], SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7) possess homology to yeast Sir2 protein [9]. The class IV HDAC (HDAC11) share sequence homology with both class I and class II HDACs [10]. Class I, II, and IV HDACs are zinc-dependent HDACs, while class III HDACs are nicotinamide adenine dinucleotide (NAD+)-dependent HDACs. HDACs are known to play important roles in various chronic diseases, such as diabetes, cardiovascular diseases, and neurodegenerative diseases [11-13].
- There has been increasing evidence that HDAC4 is involved in inflammatory and metabolic processes [14-16]. HDAC4 is ubiquitously expressed in the brain, heart, liver, skeletal muscle, and macrophages [4]. The global deletion of Hdac4 in mice is lethal, so cell-specific knockout mice have been used to dissect the roles of HDAC4 in various cell types [17-19]. HDAC4 expression and activity can be regulated at transcription, post-transcription, and post-translation [8,20,21]. In particular, posttranslational modifications in HDAC4 impact its interaction with other proteins, modulating metabolic and inflammatory processes. This review summarizes the current knowledge of the regulation and functions of HDAC4 in inflammation, insulin signaling and glucose metabolism, and cardiac muscle development.
INTRODUCTION
- The N-terminal domain of HDAC4 contains a nuclear localization signal responsible for its cellular distribution and another region needed for its interaction with the myocyte enhancer factor 2 (MEF2) family, referred to as MEF2-binding domain or MEF2-interacting domain (Fig. 1) [22]. The N-terminal region of HDAC4 contains a high level of glutamine that enables HDAC4 to interact with other glutamine-rich proteins to facilitate protein complex formation [23]. The C-terminal catalytic domain of HDAC4 is essential for substrate recognition, corepressor complex formation, and deacetylase activity [24,25]. HDAC4 does not have a strong deacetylase activity alone, but its C-terminal interaction with HDAC3 via nuclear receptor corepressor (N-CoR) in a corepressor complex can increase its enzyme activity [26-29].
STRUCTURE OF HDAC4
- Transcriptional regulation of HDAC4 by transcription factors, specificity protein 1 (Sp1) and Sp3
- HDAC4 is ubiquitously present in various tissues, and its expression is regulated differently depending on stimuli [1,5]. However, the mechanisms for the transcriptional regulation of HDAC4 remain elusive. Transcription factors, Sp1 and Sp3, are known to regulate HDAC4 transcription by directly binding to consensus GC-rich sequences in the HDAC4 promoter [30]. Transfection of Sp1 or Sp3 with a reporter driven by HDAC4 promoter increased reporter activity in melanogaster SL2 cells, which lack endogenous Sp1/Sp3 [30]. Also, overexpression of Sp1 and Sp3 upregulated HDAC4 protein levels, while HDAC4 was downregulated by Sp1 or Sp3 knockdown in HeLa cells, a human cancer cell line [30].
- Recent studies have revealed the interplay between SIRT1, a class III HDAC, and HDAC4. Inhibition of SIRT1 activity augmented ethanol-induced increase in Hdac4 expression and pro-inflammatory gene expression in macrophages [31]. Importantly, Sp1 binds to the SIRT1 promoter, increasing its promoter activity in Huh7 cell, human hepatoma cell line [32]. Therefore, SIRT1 may regulate HDAC4 transcription by competing for Sp1 with HDAC4. However, the question remains unanswered as to whether SIRT1 transcriptionally represses HDAC4 expression.
- Post-transcriptional regulation of HDAC4 by microRNA
- MicroRNAs (miRNAs) are non-coding RNAs that post-transcriptionally regulate gene expression. Recent studies have shown that miRNAs control HDAC4 expression. miR-140 targets HDAC4 at its 3´-untranslated region (3´-UTR) in 3T3 cells, mouse embryonic fibroblasts, to inhibit its translation [32]. Overexpression of miR-140 significantly decreased HDAC4 protein level without a considerable mRNA degradation [33], supporting that miR-140 regulates HDAC4 protein expression at the translational level. In addition, when neonatal rat cardiomyocytes were transfected with miR-22 mimics, HDAC4 mRNA and protein levels were reduced, while the miR-22 hairpin inhibitor, competing with target mRNA for programmed RNA-induced silencing complex, led to opposing effects on HDAC4 expression [33]. HDAC4 mRNA and protein levels were also increased in the heart of miR-22 global knockout mice, suggesting that HDAC4 mRNA stabilization and translation might be repressed by miR-22 [33]. Similarly, in human hepatocellular carcinoma tissues where miR-22 levels were reduced, there was a corresponding increase in the expression of HDAC4 protein [34]. Therefore, both miR-140 and miR-22 appear to decrease HDAC4 expression.
- Some miRNAs are known to destabilize HDAC4 mRNA. As miR-20a targets the 3´-UTR of HDAC4, knockdown of miR-20a increased HDAC4 protein in human mast cell line-1 (HMC1), while miR-20a overexpression decreased the protein levels [35]. It is also shown that HDAC4 is a direct target of miR-222, and therefore, overexpression of miR-222 decreases HDAC4 protein, suppressing apoptotic death in human chondrocytes [36]. Similarly, miR-200a directly targeted the 3´-UTR of the HDAC4 mRNA and repressed its expression in HepG2 and SMMC‐7721 cells, human liver cancer cells [37]. Interestingly, HDAC4 inhibited the expression of miR-200a and its promoter activity by decreasing histone H3 acetylation at the miR-200a promoter in SMMC‐7721 cells [37], suggesting that HDAC4 can regulate its own expression by repressing its negative regulator. In addition, HDAC4 and miR-138 regulate each other in lipopolysaccharide (LPS)-treated fibroblast-like synoviocytes in such a way that miR-138 decreases HDAC4 expression while HDAC4 decreases the histone H3 acetylation level, decreasing miR-138 transcription [38]. miR-19a-3p reduces the expression of HDAC4 in human mesenchymal stem cells; however, when both HDAC4 and miR-19a-3p are overexpressed simultaneously, the effect of miR-19a-3p on reducing HDAC4 levels is counteracted, reversing the function of miR-19a-3p reducing the osteogenic differentiation [39]. Taken together, several miRNAs that can bind to the 3´-UTR of HDAC4 regulate HDAC4 expression through mRNA degradation or inhibition of translation, while HDAC4 may modulate some miRNA expression by altering the acetylation state of their promoter. Future research is needed to understand how HDAC4 regulates specific miRNAs, including the precise location of HDAC4 binding sites on the miRNA promoters.
- Post-translational regulation of HDAC4
- Post-translational modifications of HDAC4, including phosphorylation, sumoylation, and carbonylation, influence its subcellular localization and interaction with other proteins [1,4,5]. Depending on the phosphorylation status, HDAC4 shuttles between the nucleus and cytoplasm [40-43]. Phosphorylated HDAC4 is retained in the cytoplasm through its binding to 14-3-3 proteins [1,4,5]. When dephosphorylated at Ser298 by protein phosphatase 2A (PP2A), HDAC4 is dissociated from 14-3-3 proteins, subsequently entering the nucleus [44]. The phosphorylation of HDAC4 at Ser265 and Ser266 by calcium/calmodulin-dependent kinase II (CaMKII) facilitates its nuclear export by dimerizing with HDAC5, diminishing the capacity of HDAC4 in repressing its target gene expression in neonatal rat ventricular myocytes [45,46]. Also, protein kinase A (PKA) phosphorylates HDAC4, inducing its proteolytic cleavage [47]. While CaMKII activation mediates cytosolic accumulation of HDAC4 and activation of MEF2-dependent gene programs, cyclic adenosine monophosphate (cAMP)-dependent PKA activation mediates proteolytic processing of HDAC4 to generate an N-terminal HDAC4 cleavage product, which acts as a CaMKII-insensitive repressor of MEF2 in neonatal rat ventricular myocytes [48]. Thus, the phosphorylation of HDAC4 by CaMKII or PKA counteracts MEF2 activity. Glycogen synthase kinase 3β (GSK3β) also phosphorylates HDAC4 at Ser298 and Ser302, leading to its proteasome-mediated degradation in NIH3T3 cells, mouse embryonic fibroblast cells [49].
- Sumoylation can alter HDAC4 protein expression, as sumoylation of HDAC4 has been associated with its nuclear retention, impacting HDAC4 activity as a transcriptional regulator. HDAC4 undergoes small ubiquitin-like modifier (SUMO) 1 modification at Lys559 in human embryonic kidney 293 cells, and it is also modified by SUMO-2 in mouse kidney tissue [50]. This modification is catalyzed by RNA binding protein 2 (RanBP2), a factor newly identified as a SUMO E3 ligase located in the nuclear pore complex [51]. Therefore, sumoylation may be an essential regulatory mechanism for transcriptional repression mediated by HDAC4.
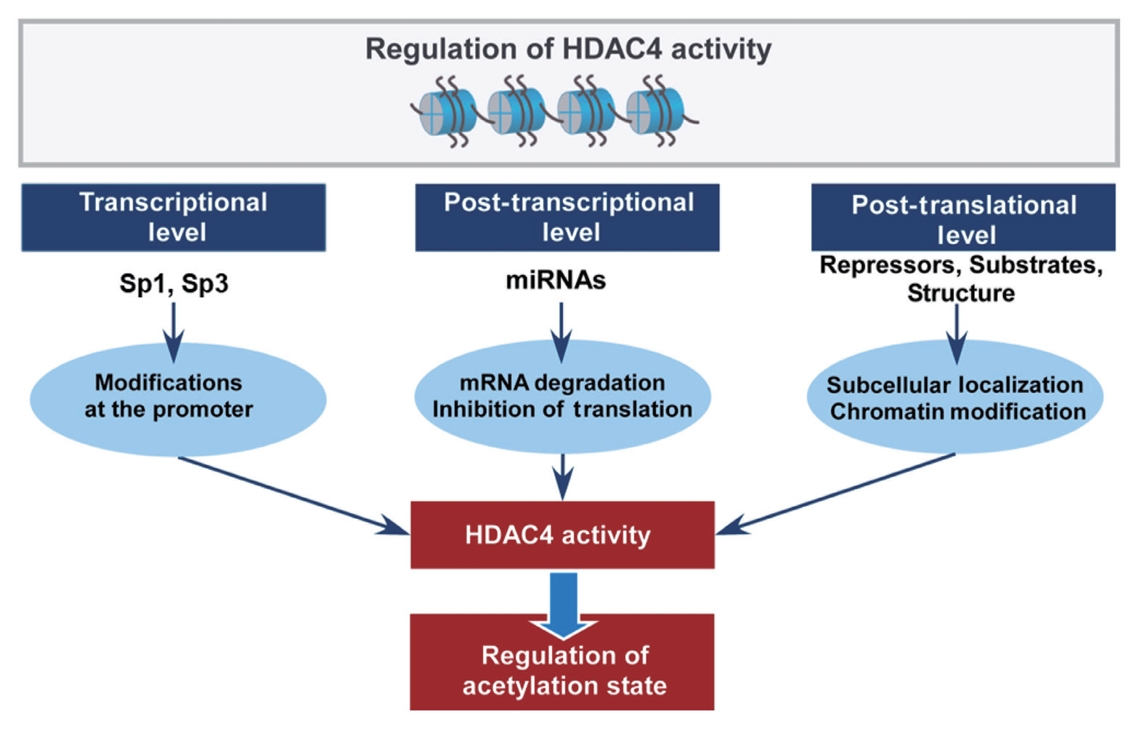
- As HDAC4 can undergo multiple post-translational modifications, there is a potential conflict between some modifications regulating HDAC4. For example, the sumoylation of HDAC4 promotes the nuclear localization of HDAC4, thus enhancing its ability; however, phosphorylation promotes the export of HDAC4 from the nucleus to the cytoplasm, repressing its ability. The regulation mechanisms for HDAC4 expression and activity are summarized in Fig. 2.
REGULATION OF HDAC4 ACTIVITY
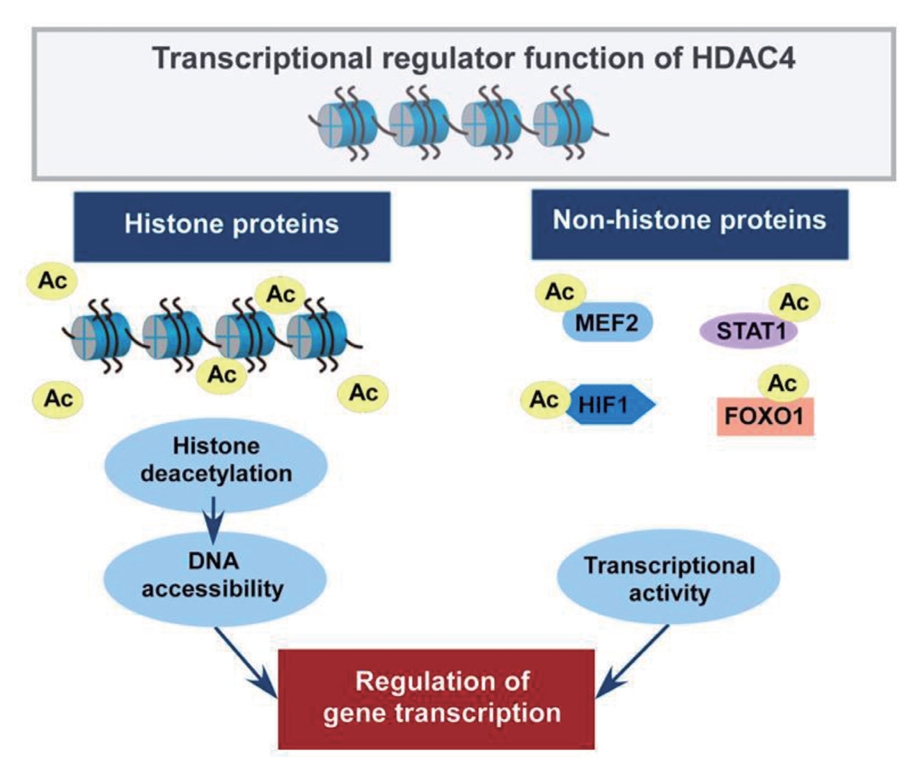
- Studies have identified that the acetylation state at the lysine residue of non-histone proteins modulates their stability, enzymatic activity, and subcellular localization [2,3]. In addition, class IIa HDACs have been reported to interact with non-histone proteins to regulate their functions [6,8]. In particular, HDAC4 plays a role in metabolic and inflammatory pathways by regulating MEF2, hypoxia-inducible factor (HIF), signal transducer and activator of transcription 1 (STAT1), and forkhead box (FOX) proteins [52-56]. Fig. 3 depicts the roles of HDAC4 in regulating histone and non-histone proteins.
- Roles of HDAC4 in the regulation of MEF2
- HDAC4 has a high binding affinity to MEF2 and functions as its repressor through chromatin remodeling via deacetylation [57-59]. The binding of HDAC4 to MEF2 facilitates the recruitment of corepressor complexes, such as silencing mediator of retinoic acid and thyroid hormone receptor (SMRT)/N-CoR-HDAC3, to repress gene expression [4,27]. As MEF2 plays a crucial role in myogenesis [60], the inhibitory effect of HDAC4 on MEF2 transcriptional activity leads to the suppression of differentiation in skeletal muscles [61,62]. Furthermore, HDAC4 represses MEF2A-mediated glucose transporter 4 (GLUT4) transcription, inhibiting glucose uptake in skeletal muscle [63]. In addition, the caspase-cleaved N-terminal fragment of HDAC4 accumulates in the nucleus, which represses the transcription of MEF2, inducing cell death [59,64].
- HDAC4 can also regulate MEF2 transcription and activity through post-transcriptional modifications, such as sumoylation and acetylation. As HDAC4 interacts with Ubc9, a SUMO E2 conjugating enzyme, and undergoes self-sumoylation, it exhibits a SUMO E3 ligase activity that allows it to repress the expression of MEF2 in 293T cells, human embryonic kidney cells [65]. As a transcriptional corepressor of MEF2, HDAC4 enhances the sumoylation of MEF2D at Ser444 in HEK293 cells. Interestingly, the sumoylation of HDAC4 itself inhibits this action [60].
- HDAC4 controls MEF2 transcriptional activity by coordinated phosphorylation and sumoylation [66]. HDAC4 recruits cyclin-dependent kinase 5 (Cdk5) to phosphorylate MEF2 at Ser444, and this phosphorylation promotes Lys439 sumoylation to decrease MEF2 transcriptional activity, as HDAC4 likely acts as a SUMO E3 ligase [66]. This phosphorylation and sumoylation of MEF2, particularly sumoylation, often represses MEF2 transcriptional activity. Interestingly, HDAC4 promotes sumoylation on a lysine residue, which is usually acetylated by a MEF2 coactivator, cAMP-response element binding protein (CBP), suggesting that HDAC4 may not function as a deacetylase for MEF2, despite its typical function as a HDAC [65]. The acetylation of MEF2 induces MEF2 activation, while sumoylation represses MEF2 transcriptional activity [60,66]. This finding indicates a possible interplay between acetylation and sumoylation in regulating MEF2 activity. Further study is necessary to clarify whether HDAC4 directly sumoylates MEF2 or recruits Ubc9 for MEF2 sumoylation.
- Roles of HDAC4 in the regulation of HIF
- HIF1, a transcription factor crucial for adaptive cellular response to hypoxia, plays an essential role in the transcriptional regulation of genes involved in various biological processes, including glucose metabolism, angiogenesis, and cell survival [67]. In response to hypoxia, often due to solid tumors and ischemic lesions, stressed cells increase HIF1 expression to upregulate vascular endothelial growth factor (VEGF) and other angiogenic factors in cancer [68]. HDAC4 enhances HIF1α transcriptional activity by binding to HIF1α inhibitory domain [69], which is also the binding site for factor inhibiting HIF1 (FIH-1) that typically inhibits HIF1α transcriptional activity [69]. By binding to this site, HDAC4 can prevent the inhibitory action of FIH-1 on HIF1α, thus enhancing HIF1α transcriptional activity in HepG2 cells [70]. In addition, HDAC4 increases the transactivation function of HIF1α, but it does this not by interacting with FIH-1, but rather by binding to p300, a transcriptional coactivator [7]. Besides, HDAC4 increased VEGF expression without increasing HIF1α transcription or protein stability, indicating HDAC4 is likely to increase HIF1α transcriptional activity at the post-translational level in HepG2 cells [70].
- HIF1 is deacetylated at several lysine residues by several HDACs [71]. HDAC4 positively regulates the HIF1α activity in hypoxic conditions, allowing the cell to respond effectively to low oxygen levels [7]. In von Hippel-Lindau-deficient human renal carcinoma cell line, UMRC2, HDAC4 knockdown or HDAC inhibitors reduced HIF1α protein and transcriptional activity [72]. Also, when HDAC4 was knocked down, HIF1α acetylation was increased concomitantly with decreased stability, which was reversed by HDAC4 overexpression in Hep3B cells, human liver cancer cells [73]. It is known that HDAC4 stabilizes HIF1α by inducing the interaction between HIF1α and heat shock protein 90kD (Hsp90) [73], as Hsp90 helps HIF1α complete its maturation under stressful conditions [74]. Thus, HDAC4 increases the stability of HIF1α and its transcriptional activity by several mechanisms.
- Role of HDAC4 in the regulation of STAT1
- STAT1, one of the seven mammalian STAT families, is vital in innate and adaptive immune systems [75]. The activation of STAT1 is regulated by both phosphorylation and acetylation [75]. Pan-HDAC inhibitors, such as trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), enhanced STAT1 acetylation at Lys410 and Lys413, increasing STAT1 mRNA and protein levels in SK-37 and NW-1539 human melanoma cells [76]. The increased acetylation of STAT1 by TSA inhibits interferon (IFN)-dependent STAT1 phosphorylation, which is required for STAT1 activation. Therefore, this inhibition decreases the nuclear translocation of STAT1, lowering its transcriptional activity in human embryonic kidney 293T cells [75]. Also, in human lung cancer A549 cells, elevated STAT1 acetylation by TSA reduced its transcriptional activity, leading to decreased cell migration, invasion, and sphere formation [77].
- When HDAC4 was knocked down, STAT1 transcriptional activity was inhibited, repressing cell migration and invasion, indicating that STAT1 is an HDAC4 target [7]. TSA treatment reduced HDAC4 expression along with phosphorylated STAT1, and STAT1 knockdown reduced the HDAC4 expression in etoposide-resistant A549 cells [78]. Also, silencing HDAC4 increased acetylated STAT1 levels, preventing platinum-induced STAT1 activation, while HDAC4 overexpression promotes STAT1 deacetylation and cancer cell survival in response to platinum exposure in ovarian cancer cells [79]. HDAC4 also regulates the acetylation and phosphorylation of STAT1 in LPS-stimulated RAW 264.7 macrophages, which requires the acetylation of high mobility group box 1 (HMGB1) [80]. In particular, when HDAC4’s degradation is inhibited by treatment with a toll-like receptor (TLR) inhibitor and a JAK inhibitor, the acetylation of HMGB1 is decreased, leading to reduced STAT1 phosphorylation in LPS-stimulated RAW 264.7 macrophages [80].
- HDAC4 suppresses inflammatory responses mediated by STAT1 by facilitating its sumoylation, which involves the liver X receptor (LXR) in IFNγ-stimulated brain astrocytes [81]. As HDAC4 acts as a SUMO E3 ligase [60], LXRα formed a complex with HDAC4 and phosphorylated STAT1. Taken together, HDAC4 plays a crucial role in regulating STAT1 through two primary mechanisms: the activation by deacetylation and the inhibitions of STAT1 activity through sumoylation.
- Roles of HDAC4 in the regulation of forkhead box O1
- Forkhead box O (FOXO) proteins belong to class O of the forkhead family and have essential roles in glucose and lipid metabolism, cell proliferation, stress resistance, and apoptosis [82]. In particular, FOXO1 is sensitive to insulin to regulate gene transcription [83].
- A crucial role of HDACs in the pathogenesis of diabetes mellitus is well known, as two pan-HDAC inhibitors, i.e., TSA and sodium butyrate, stimulate the transcription of insulin-sensitive genes, such as glucose 6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK), by increasing histone H4 acetylation [84]. This result suggests that HDACs may regulate FOXO1. Also, MC1568, a class IIa HDAC-specific inhibitor, and pan-HDAC inhibitors decrease FOXO1 enrichment at the cis-element of target gene promoters in mouse insulinoma 6 cells treated with high glucose [84]. Pan-HDAC inhibitors, including sodium valproate (VPA), TSA, and SAHA, repress the transcriptional activity of FOXO1 in HepG2 cells [85]. Importantly, HDAC4 knockdown increased FOXO1 acetylation, decreasing its activity and gluconeogenic gene expression [85]. However, HDAC4 knockdown results in an increase in FOXO1 and gluconeogenic gene expression, such as G6Pase and PEPCK, as well as a decrease in phosphorylated FOXO1, further contributing to increased gluconeogenesis in HepG2 cells [86]. The reason for the contradictory findings between the two studies is unclear. During fasting, glucagon leads to the dephosphorylation and nuclear entry of HDAC4, and then nuclear HDAC4 interacts with HDAC3, promoting the activation of gluconeogenic genes via deacetylation and activation of FOXO in mouse primary hepatocytes [26]. However, the knockdown of HDAC4 in mouse liver increased FOXO acetylation and inhibited FOXO target genes, consequently lowering blood glucose [26]. When cells were exposed to diabetes‐associated HDAC4 mutations, including p.His227Arg, p.Asp234Asn, and p.Glu374Lys, HDAC4 mutations export FOXO1 from the nucleus to the cytosol by increasing its acetylation levels in Min6 cells, a mouse pancreatic β-cell line, reducing insulin secretion [87].
- Salt-inducible kinase 3 (SIK3) and HDAC4 are known to regulate FOXO activity in drosophila and HeLa cells [88]. During feeding, active SIK3 phosphorylates HDAC4, blocking FOXO deacetylation; however, during fasting, HDAC4 is dephosphorylated due to inactive SIK3, enabling FOXO deacetylation [88]. Furthermore, inhibiting class IIa HDACs, either by treatment with TSA, or by knockdown of HDAC4/5, interfered with the expression of gluconeogenic FOXO target genes in HepG2 cells [88].
- Taken together, when HDAC4 is dephosphorylated and translocated into the nucleus, it directly interacts with FOXO1. Deacetylation of FOXO1 by HDAC4 can enhance its transcriptional activity.
ROLES OF HDAC4 IN THE REGULATION OF NON-HISTONE PROTEINS
- HDAC4 regulates inflammatory and metabolic processes by deacetylating various transcription factors crucial for those pathways. Thus, this section summarizes the effects of HDAC4 on inflammation, insulin signaling and glucose metabolism, and cardiac muscle development.
- Roles of HDAC4 in inflammation
- The knockdown of HDAC4 decreased the pro-inflammatory gene expression in LPS-treated RAW 264.7 macrophages [14]. Consistently, HDAC4 knockdown decreased the pro-inflammatory gene expression, while overexpression of HDAC4 further increased the pro-inflammatory gene expression in ethanol-stimulated RAW 264.7 macrophages [31]. Pro-inflammatory cytokines produced by LPS-stimulated macrophages require HDAC4, facilitating the metabolic switch to aerobic glycolysis, which is controlled by GSK3β and inducible nitric oxide synthase [89]. Interestingly, glycolysis activates caspase-3, which cleaves HDAC4 for degradation, limiting HDAC4-dependent inflammation [89].
- Studies have shown that HDAC4 can increase the activity of transcription factors involved in the inflammatory signaling pathways, such as nuclear factor κB (NFκB) and activator protein 1 (AP1). HDAC4 induces vascular inflammation by increasing NFκB transcriptional activity and reactive oxygen species production in tumor necrosis factor α (TNFα)-stimulated vascular smooth muscle cells [90]. In addition, HDAC4 siRNA inhibits TNF-induced phosphorylation of NFκB p65 in cultured rat mesenteric arterial smooth muscle cells, decreasing pro-inflammatory mediators [91]. However, it remains to be seen how HDAC4 regulates p65 phosphorylation. HDAC4 also stimulates AP1 activity to induce pro-inflammatory gene expression by promoting deacetylation, activating a mitogen-activated protein (MAP) kinase kinase kinase 2 (MEKK2), in mouse-denervated muscle [92].
- It is of interest that studies have also shown the inhibitory effects of HDAC4 on inflammatory pathways. HDAC4 mediates inflammatory pathways by interacting with pattern recognition receptors (PRRs) that respond to external stimuli [93]. The overexpression of HDAC4 suppresses the import of production of type I IFNs triggered by PRRs, preventing an over-reactive innate immune response in HEK-293 cells [93]. TLRs are well-known PRRs interacting with HDAC4 [94]. As NFκB becomes activated in response to TLR signaling to induce proinflammatory cytokines [94-96], HDAC4 may link with regulating inflammatory responses mediated by TLR signaling and NFκB activation. The overexpression of HDAC4 significantly impaired TNFα-dependent activation of NFκB in HEK-293, human embryonic kidney cells, indicating a protective role of HDAC4 against NFκB activity to regulate immune response [97]. Also, cAMP stimulates HDAC4 activity to repress inflammation by activating PKA, inhibiting the SIKs [15,98]. As SIKs phosphorylate HDAC4 for its cytoplasmic sequestration, inhibition of SIKs enables dephosphorylated HDAC4 to shuttle to the nucleus, inhibiting NFκB activity [15]. Recently, it has been reported that the cytoplasmic location of HDAC4 is essential for inhibitor kappa B-alpha (IκBα) sumoylation leading to the inhibition of TNFα- or LPS-triggered NFκB activation in HEK293T cells [99]. Therefore, the role of HDAC4 in the regulation of inflammation may differ depending on extracellular stimuli and types of cells, which needs further investigation.
- Studies have shown conflicting observations as to the effects of HDAC4 on NFκB activity, as described above. miRNAs modulate HDAC4 expression by mRNA degradation or inhibition of translation, while HDAC4 can regulate miRNA expression by altering the acetylation level of their promoter. miR-20a suppresses HDAC4 and inhibits the expression of pro-inflammatory cytokines, including TNFα, interleukin 1β (IL-1β), and IFNγ, but promotes IL-10 production, attenuating allergic inflammation in HMC-1 cells [35]. As IL-10 inhibits TNFα production by inhibiting NFκB activation [100], HDAC4 might be involved indirectly in inducing inflammatory cytokines, possibly through a miR 20a-HDAC4-IL-10 pathway in human mast cells, which may be associated with chronic inflammatory disease of lung airways [35,100]. As miR-138 directly targets HDAC4 to activate NFκB signaling to promote rheumatoid arthritis, HDAC4 deficiency further increases miR-138-induced expression of inflammatory cytokines, such as IL-6, IL-1β, TNFα, and IL-8 [38,72]. As conflicting evidence exists regarding the regulatory role of HDAC4 in modulating NFκB activity, the interaction between HDAC4 and miRNAs and the subsequent effects on inflammation are complicated. These observations highlight the intricate and context-dependent roles of HDAC4 in modulating inflammatory responses, which requires further study to fully elucidate its function in the inflammatory pathways.
- Roles of HDAC4 in the insulin signaling and glucose metabolism
- HDAC4 plays a role in metabolic pathways. The osteoblast-specific deletion of HDAC4 was associated with the loss of weight and appetite in rodents [5]. Macrophage-specific deletion of HDAC4 exhibited higher blood glucose and free fatty acid concentrations with glucose intolerance after 8 weeks on a high-fat diet compared with control mice, indicating HDAC4 deficiency in macrophages promoted insulin resistance and obesity [15]. In human peripheral blood mononuclear cells, HDAC4 was downregulated in obese subjects, which was increased by physical exercise [97]. Also, evaluation of the association between HDAC4 variants and obesity indicates that the cAMP-HDAC4 pathway is essential in maintaining insulin sensitivity and energy balance by modulating the innate immune system [63].
- The role of HDAC4 in insulin signaling has been reported. When diet-induced obesity mice were injected with an adenoviral vector encoding mutant HDAC4 constitutively expressed in the nucleus, the insulin signal was improved in the hepatocyte, and hyperglycemia and hyperinsulinemia were attenuated [101]. While HDAC4 in osteoblasts is shown to increase appetite [102], HDAC4 deficiency in osteoblasts exhibited decreased circulating and pancreatic insulin levels [5]. This result indicates that HDAC4 in osteoblasts is required for insulin production or secretion. Treatment of adenoviral vectors encoding mutant Hdac4, which are liver-targeted, phosphorylation-defective to enforce nuclear HDAC4, improved the insulin signaling in the hepatocyte and protected against hyperglycemia and hyperinsulinemia in diet-induced obesity mice [101]. HDAC4 can induce gluconeogenic enzymes through deacetylation and activation of FOXO, while its deficiency increases FOXO acetylation for inactivation in hepatocytes and mouse liver, lowering blood glucose in mice fed a high-fat diet [26]. Also, overexpression of HDAC4 reduced insulin-secreting β-cell mass with decreased insulin mRNA expression in the pancreas during embryonic development [103]. However, the osteoblast-specific HDAC4 knockout mice showed decreased β-cell area and insulin contents in the pancreas with reduced insulin secretion [5]. It is unclear why the inconsistency exists regarding the role of HDAC4 in insulin production and secretion, requiring further investigation.
- Abnormal glucose metabolism is a hallmark of insulin resistance and type 2 diabetes mellitus. Under the fasting condition where serum glucagon is high, HDAC4 is dephosphorylated, translocated into the nucleus, and recruited to the promoter of gluconeogenic G6Pase for its induction [26]. Knocking down HDAC4/5/7 significantly improved glucose intolerance in leptin-deficient mice [26]. As HDAC4/5 interacts directly with HDAC3, resulting in the deacetylation and activation of FOXO family transcription factors [26], the loss of HDAC4 in the murine liver may lead to reductions in FOXO target gene expression, consequently lowering blood glucose.
- HDAC4 can also disturb glucose metabolism by inhibiting the transcription of GLUT4. HDAC4 deficiency increased GLUT4 protein in human 3T3L1 adipocytes [104]. In response to fasting, HDAC4/5 represses the transcription of GLUT4 by binding to its promoter in white adipose tissue [105] and C2C12 myotubes [106]. Physical exercise decreased HDAC4/5 activity in the skeletal muscle of C57BL/6J mice, accompanied by increased GLUT4 transcription [106]. Thus, HDAC4 upregulates gluconeogenic genes but downregulates GLUT4 transcription. Interestingly, HDAC4 is reported to increase the transcriptional activity of HIF1α by enhancing its dissociation from its inhibiting factor, asparaginyl hydroxylase, in HepG2 cells [70] and knockdown of Hdac4 decreased the stability of HIF1α protein in Hep3B cells, human liver cancer cells [73]. Similarly, protein levels and transcriptional activity of HIF1α were also reduced by HDAC4 knockdown in a human renal cell carcinoma cell line [72]. As HDAC4 deficiency abrogated the ethanol-induced increase in glycolysis-related genes, i.e., Hif1a and Glut1, in bone marrow-derived macrophages [31], HDAC4 may play a role in the metabolic shift to favor glycolysis. Collectively, HDAC4 can be a potential target for the improvement of glucose homeostasis.
- Roles of HDAC4 in cardiac muscle development
- As HDAC4 directly binds to MEF2 for repression [60], HDAC4 may affect cardiac muscle development. In particular, HDAC4 plays an inhibitory role in cardiac hypertrophy [1] and regulates the differentiation of mesoderm cells into cardiomyoblasts [4,5]. Overexpression of HDAC4 inhibits cardiomyogenesis and downregulates cardiac muscle gene expression during the differentiation of P19 embryonic carcinoma stem cells into cardiomyocytes [107]. It has been reported that MEF2C knockout mice exhibit defective heart morphogenesis, as they are arrested at the heart looping stage, which is crucial for the maturation of the heart [108]. Since HDAC4 is a negative regulator for MEF2, it is presumable that HDAC4 may induce a similar phenotype to MEF2C knockout.
- In addition, myocyte-specific overexpression of HDAC4 decreases myocardial function and increases myocardial ischemia/reperfusion injury, which is associated with increased autophagy and apoptosis in the heart [109]. However, the silencing of HDAC4 rescues Neat1-induced cardiomyocyte apoptosis in diabetic cardiomyopathy mice, resulting in cardioprotection against diabetic cardiomyopathy [110]. Taken together, HDAC4 may play an inhibitory role in cardiac muscle development.
ROLES OF HDAC4 IN THE INFLAMMATORY AND METABOLIC PROCESSES
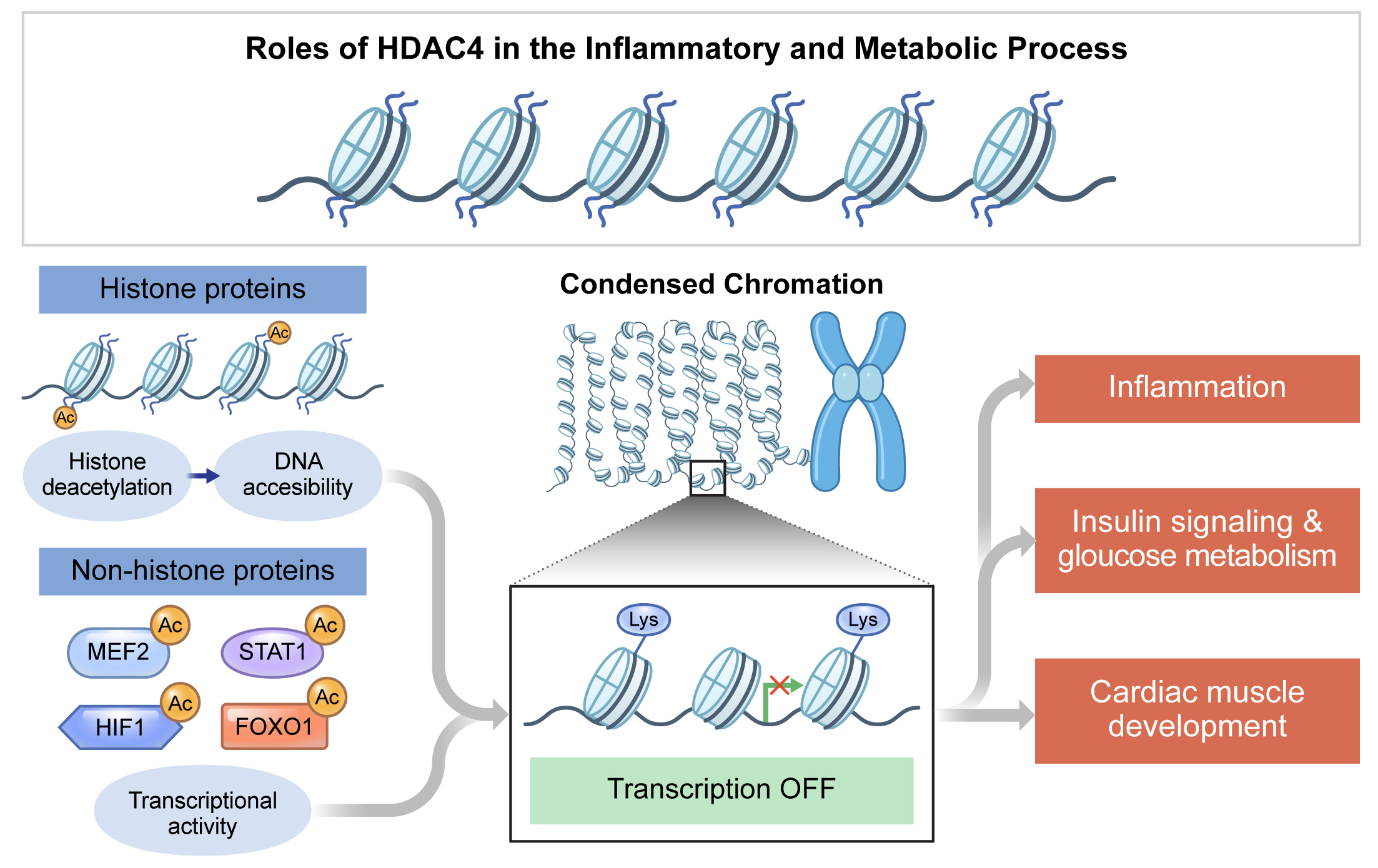
- The roles of HDAC4 in the inflammatory and metabolic processes are summarized in Table 1. HDAC4 is regulated by various endogenous and exogenous factors and its substrates, especially non-histone targets, including transcription factors (summarized in Figs. 2 and 3). The complicated, inconsistent, and even somewhat opposing functions of HDAC4 in different cell types and tissues may be partly due to its structure, cellular localization, and interaction with different substrates. Further investigations of downstream and upstream targets of HDAC4 will extend our knowledge of the specific biological roles of HDAC4, leading to the development of therapeutic strategies for inflammatory and metabolic processes.
CONCLUSIONS
-
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
-
FUNDING
None
NOTES
-
Acknowledgements
- No potential conflict of interest relevant to this article was reported.
Fig. 1.The histone deacetylase (HDAC) superfamily. Green bars indicate the zinc-dependent deacetylase domain; Blue bars indicate the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase domain; Yellow bars indicate the myocyte enhancer factor 2 (MEF2) binding domain; Pink bars indicate the nuclear localization signal; Brown bars indicate the nuclear export signal; Purple bar indicates the zinc finger domain. SIRT, sirtuin.
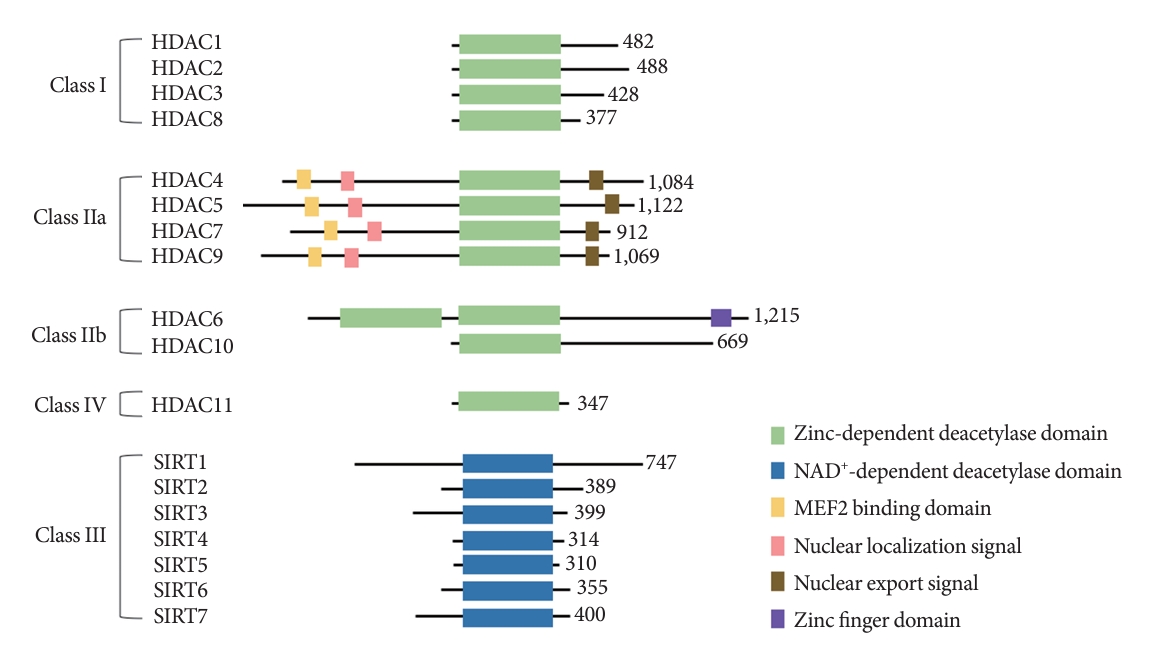

Fig. 2.Summary of the regulatory modes of histone deacetylase 4 (HDAC4) expression and activity. The transcriptional level of HDAC4 is regulated by transcription factors, specificity protein 1 (Sp1) and Sp3, by directly binding to consensus GC-rich sequences in the HDAC4 promoter. miRNAs regulate HDAC4 expression at the post-transcriptional level of HDAC4 through mRNA degradation or inhibition of translation, while HDAC4 can also regulate the expression of miRNAs by altering the acetylation states of their promoter. The post-translational level of HDAC4 regulation includes its subcellular localization and interaction with other proteins as repressors, substrates, or structures. As HDAC4 undergoes several post-translational modifications, there might conflict with each other for the regulation of HDAC4, which influence the localization of HDAC4. The modifications in HDAC4 are involved in a wide range of biological processes by regulating the acetylation state.
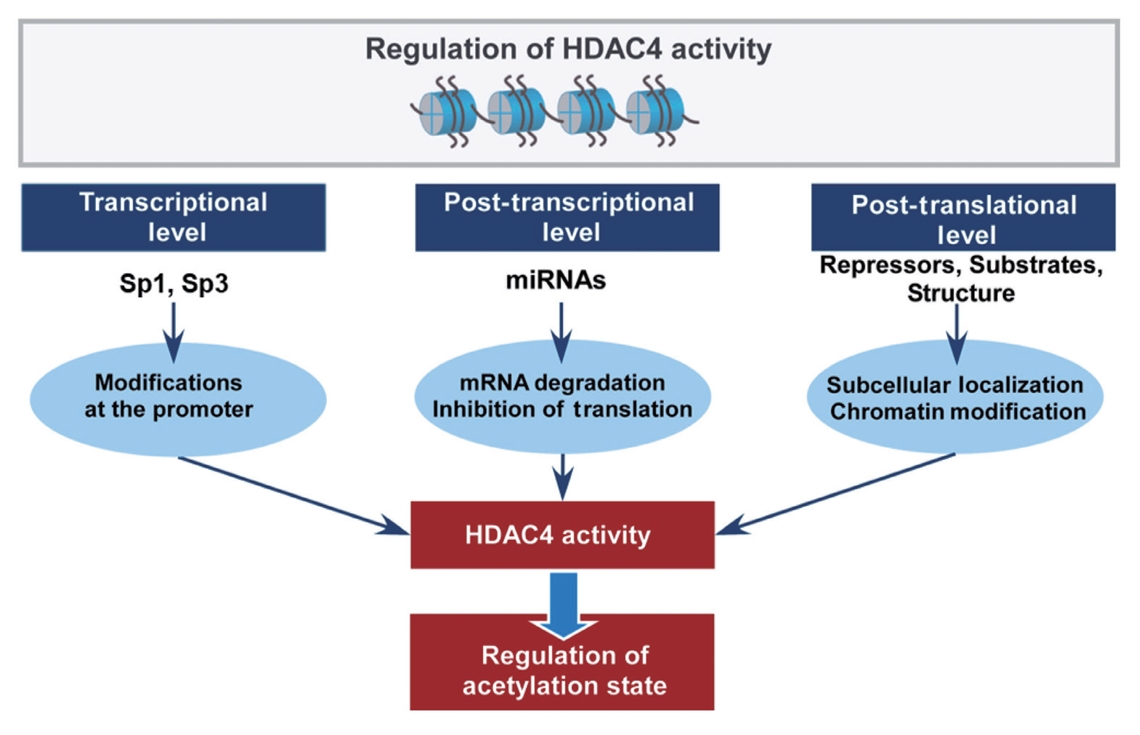

Fig. 3.The role of histone deacetylase 4 (HDAC4) in the regulation of histone and non-histone proteins. The acetylation state at the lysine residue regulates the function of histone and non-histone proteins. Histone deacetylation by HDAC4 condenses chromatin structure, which controls the binding of transcription factors to DNA for transcriptional repression. HDAC4 interacts with non-histone proteins, including transcription factors, as a transcriptional corepressor. In particular, HDAC4 plays a role in metabolic and inflammatory pathways by regulating myocyte enhancer factor 2 (MEF2), hypoxia-inducible factor 1 (HIF1), signal transducer and activator of transcription 1 (STAT1), and forkhead box O 1 (FOXO1) transcription factors. The modifications of histone and non-histone proteins are involved in a wide range of biological processes by regulating gene transcription.
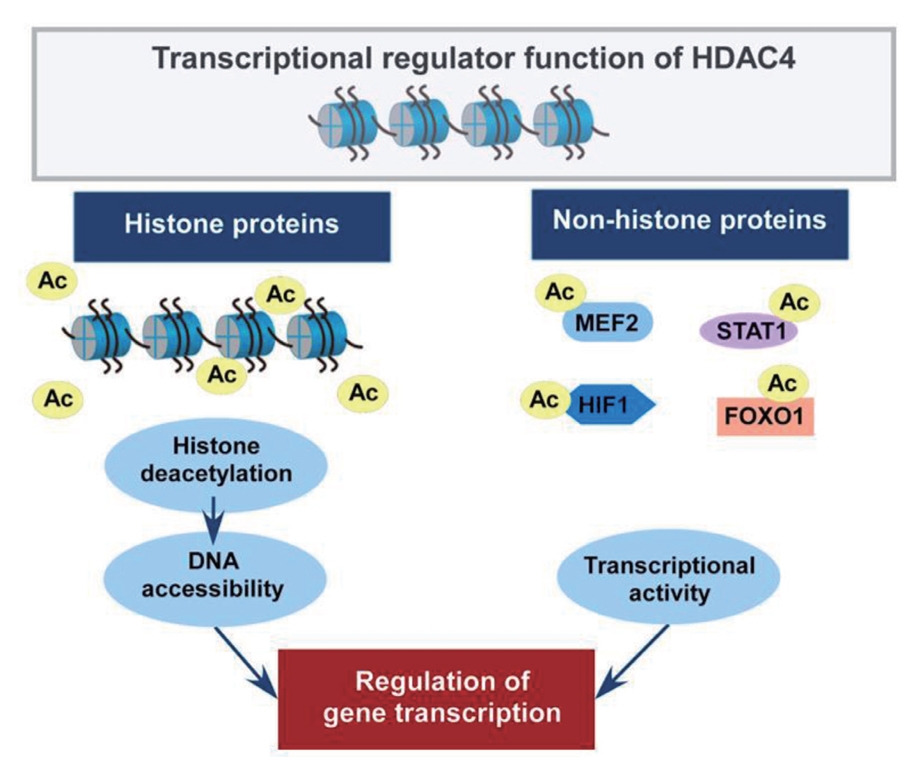

Table 1.Summary for the regulatory actions of HDAC4
| Response or signaling | HDAC4 presence | Regulation | Change | Reference |
|---|---|---|---|---|
| Inflammation | HDAC4 presence | NFκB transcriptional activity and ROS production in TNFα-stimulated rat primary vascular smooth muscle cells | ↑ | [90] |
| AP1 activity in mouse denervated muscle | ↑ | [92] | ||
| Pro-inflammatory cytokines in LPS-stimulated BV2 cells | ↑ | [89] | ||
| Type I IFNs secretion in HEK-293 cells | ↓ | [93] | ||
| NFκB transcriptional activity in TNFα- or LPS-stimulated HEK-293 cells | ↓ | [97,99] | ||
| NFκB transcriptional activity in LPS-stimulated human primary fibroblast-like synoviocytes | ↓ | [38] | ||
| HDAC4 absence | Pro-inflammatory gene expression in LPS-stimulated RAW 264.7 cells | ↓ | [14] | |
| Pro-inflammatory gene expression in ethanol-stimulated RAW 264.7 cells | ↓ | [31] | ||
| Insulin signaling & glucose metabolism | HDAC4 presence | Gluconeogenic enzymes and FOXO activity in glucagon-treated mouse primary hepatocytes | ↑ | [26] |
| Insulin-secreting β-cell mass in rat pancreas | ↓ | [103] | ||
| GLUT4 transcriptional activity in 3T3L1 cells | ↓ | [105] | ||
| GLUT4 transcriptional activity in C2C12 cells | ↓ | [106] | ||
| β-Cell area, insulin contents, and insulin secretion in mouse pancreas | ↓ | [5] | ||
| HDAC4 absence | Blood glucose, free fatty acid, and glucose intolerance in mouse after high-fat diet | ↑ | [15] | |
| GLUT4 protein levels in 3T3L1 cells | ↑ | [104] | ||
| HIF1α protein stability in Hep3B cells | ↓ | [73] | ||
| HIF1α protein and transcriptional activity in UMRC2 cells | ↓ | [72] | ||
| Glycolysis-related genes in ethanol-stimulated mouse primary bone marrow-derived macrophages | ↓ | [31] | ||
| Cardiac muscle development | HDAC4 presence | Cardiomyogenesis and cardiac muscle gene expression in P19 cells | ↓ | [107] |
| Myocardial ischemia/reperfusion injury in mouse heart | ↑ | [109] | ||
| HDAC4 absence | Apoptosis in mouse primary cardiomyocytes | ↓ | [110] |
- 1. Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics 2014;6:139-50.ArticlePubMed
- 2. Jenuwein T, Allis CD. Translating the histone code. Science 2001;293:1074-80.ArticlePubMed
- 3. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95.ArticlePubMedPMCPDF
- 4. Mielcarek M, Zielonka D, Carnemolla A, Marcinkowski JT, Guidez F. HDAC4 as a potential therapeutic target in neurodegenerative diseases: a summary of recent achievements. Front Cell Neurosci 2015;9:42.ArticlePubMedPMC
- 5. Makinistoglu MP, Karsenty G. The class II histone deacetylase HDAC4 regulates cognitive, metabolic and endocrine functions through its expression in osteoblasts. Mol Metab 2014;4:64-9.ArticlePubMedPMC
- 6. Peng L, Seto E. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb Exp Pharmacol 2011;206:39-56.ArticlePubMed
- 7. Clocchiatti A, Di Giorgio E, Demarchi F, Brancolini C. Beside the MEF2 axis: unconventional functions of HDAC4. Cell Signal 2013;25:269-76.ArticlePubMed
- 8. Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol 2001;79:243-52.ArticlePubMed
- 9. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713.ArticlePubMedPMC
- 10. Chen H, Xie C, Chen Q, Zhuang S. HDAC11, an emerging therapeutic target for metabolic disorders. Front Endocrinol (Lausanne) 2022;13:989305.ArticlePubMedPMC
- 11. Makkar R, Behl T, Arora S. Role of HDAC inhibitors in diabetes mellitus. Curr Res Transl Med 2020;68:45-50.ArticlePubMed
- 12. Bagchi RA, Weeks KL. Histone deacetylases in cardiovascular and metabolic diseases. J Mol Cell Cardiol 2019;130:151-9.ArticlePubMed
- 13. Shukla S, Tekwani BL. Histone deacetylases inhibitors in neurodegenerative diseases, neuroprotection and neuronal differentiation. Front Pharmacol 2020;11:537.ArticlePubMedPMC
- 14. Pham TX, Park YK, Lee JY. Anti-inflammatory effects of spirulina platensis extract via the modulation of histone deacetylases. Nutrients 2016;8:381.ArticlePubMedPMC
- 15. Luan B, Goodarzi MO, Phillips NG, Guo X, Chen YD, Yao J, et al. Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab 2014;19:1058-65.ArticlePubMedPMC
- 16. Du J, Zhang L, Wang H, Zhao YT, Yano N, Zhao TC. Cardiac specific overexpression of histone deacetylase 4 aggravates high fat diet-induced cardiac dysfunction and metabolic disorders. Circulation 2016;134(Suppl 1):A17970.
- 17. Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science 2009;323:256-9.ArticlePubMedPMC
- 18. Blixt NC, Faulkner BK, Astleford K, Lelich R, Schering J, Spencer E, et al. Class II and IV HDACs function as inhibitors of osteoclast differentiation. PLoS One 2017;12:e0185441.ArticlePubMedPMC
- 19. Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med 2012;18:783-90.ArticlePubMedPMCPDF
- 20. Luo L, Martin SC, Parkington J, Cadena SM, Zhu J, Ibebunjo C, et al. HDAC4 controls muscle homeostasis through deacetylation of myosin heavy chain, PGC-1α, and Hsc70. Cell Rep 2019;29:749-63.ArticlePubMed
- 21. Fitzsimons HL. The class IIa histone deacetylase HDAC4 and neuronal function: nuclear nuisance and cytoplasmic stalwart? Neurobiol Learn Mem 2015;123:149-58.ArticlePubMed
- 22. Borghi S, Molinari S, Razzini G, Parise F, Battini R, Ferrari S. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J Cell Sci 2001;114(Pt 24):4477-83.ArticlePubMedPDF
- 23. Guo L, Han A, Bates DL, Cao J, Chen L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci U S A 2007;104:4297-302.ArticlePubMedPMC
- 24. Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem 2008;283:26694-704.ArticlePubMedPMC
- 25. Porter NJ, Christianson DW. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr Opin Struct Biol 2019;59:9-18.ArticlePubMedPMC
- 26. Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011;145:607-21.ArticlePubMedPMC
- 27. Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 2001;21:6091-101.ArticlePubMedPMCPDF
- 28. Lee HA, Song MJ, Seok YM, Kang SH, Kim SY, Kim I. Histone deacetylase 3 and 4 complex stimulates the transcriptional activity of the mineralocorticoid receptor. PLoS One 2015;10:e0136801.ArticlePubMedPMC
- 29. Ginnan R, Sun LY, Schwarz JJ, Singer HA. MEF2 is regulated by CaMKIIδ2 and a HDAC4-HDAC5 heterodimer in vascular smooth muscle cells. Biochem J 2012;444:105-14.ArticlePubMedPDF
- 30. Liu F, Pore N, Kim M, Voong KR, Dowling M, Maity A, et al. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol Biol Cell 2006;17:585-97.ArticlePubMedPMC
- 31. Kang H, Park YK, Lee JY. Inhibition of alcohol-induced inflammation and oxidative stress by astaxanthin is mediated by its opposite actions in the regulation of sirtuin 1 and histone deacetylase 4 in macrophages. Biochim Biophys Acta Mol Cell Biol Lipids 2021;1866:158838.ArticlePubMed
- 32. Okazaki M, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, Nakayama S, et al. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocr J 2010;57:403-13.ArticlePubMed
- 33. Huang ZP, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res 2013;112:1234-43.ArticlePubMedPMC
- 34. Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. MicroRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer 2010;103:1215-20.ArticlePubMedPMCPDF
- 35. Lu Y, Li Z, Xie B, Song Y, Ye X, Liu P. hsa-miR-20a-5p attenuates allergic inflammation in HMC-1 cells by targeting HDAC4. Mol Immunol 2019;107:84-90.ArticlePubMed
- 36. Song J, Jin EH, Kim D, Kim KY, Chun CH, Jin EJ. MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis. BBA Clin 2014;3:79-89.ArticlePubMedPMC
- 37. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011;54:2025-35.ArticlePubMed
- 38. Shao L, Hou C. miR-138 activates NF-κB signaling and PGRN to promote rheumatoid arthritis via regulating HDAC4. Biochem Biophys Res Commun 2019;519:166-71.ArticlePubMed
- 39. Chen R, Qiu H, Tong Y, Liao F, Hu X, Qiu Y, et al. MiRNA19a-3p alleviates the progression of osteoporosis by targeting HDAC4 to promote the osteogenic differentiation of hMSCs. Biochem Biophys Res Commun 2019;516:666-72.ArticlePubMed
- 40. Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, Inuabasi L, et al. HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol 2013;11:e1001717.ArticlePubMedPMC
- 41. Nishino TG, Miyazaki M, Hoshino H, Miwa Y, Horinouchi S, Yoshida M. 14-3-3 regulates the nuclear import of class IIa histone deacetylases. Biochem Biophys Res Commun 2008;377:852-6.ArticlePubMed
- 42. Wu Q, Yang X, Zhang L, Zhang Y, Feng L. Nuclear accumulation of histone deacetylase 4 (HDAC4) exerts neurotoxicity in models of Parkinson’s disease. Mol Neurobiol 2017;54:6970-83.ArticlePubMedPDF
- 43. Litke C, Bading H, Mauceri D. Histone deacetylase 4 shapes neuronal morphology via a mechanism involving regulation of expression of vascular endothelial growth factor D. J Biol Chem 2018;293:8196-207.ArticlePubMedPMC
- 44. Paroni G, Cernotta N, Dello Russo C, Gallinari P, Pallaoro M, Foti C, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell 2008;19:655-67.ArticlePubMedPMC
- 45. Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest 2006;116:1853-64.ArticlePubMedPMC
- 46. Kreusser MM, Backs J. Integrated mechanisms of CaMKII-dependent ventricular remodeling. Front Pharmacol 2014;5:36.ArticlePubMedPMC
- 47. Liu Y, Schneider MF. Opposing HDAC4 nuclear fluxes due to phosphorylation by β-adrenergic activated protein kinase A or by activity or Epac activated CaMKII in skeletal muscle fibres. J Physiol 2013;591:3605-23.ArticlePubMedPMC
- 48. Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, et al. Selective repression of MEF2 activity by PKAdependent proteolysis of HDAC4. J Cell Biol 2011;195:403-15.ArticlePubMedPMCPDF
- 49. Cernotta N, Clocchiatti A, Florean C, Brancolini C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell 2011;22:278-89.ArticlePubMedPMC
- 50. Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 2001;276:35368-74.ArticlePubMed
- 51. Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 2002;21:2682-91.ArticlePubMedPMC
- 52. Kong Q, Hao Y, Li X, Wang X, Ji B, Wu Y. HDAC4 in ischemic stroke: mechanisms and therapeutic potential. Clin Epigenetics 2018;10:117.ArticlePubMedPMCPDF
- 53. Zhang P, Sun Q, Zhao C, Ling S, Li Q, Chang YZ, et al. HDAC4 protects cells from ER stress induced apoptosis through interaction with ATF4. Cell Signal 2014;26:556-63.ArticlePubMed
- 54. Sando R 3rd, Gounko N, Pieraut S, Liao L, Yates J 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 2012;151:821-34.ArticlePubMedPMC
- 55. Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 2013;14:347-59.ArticlePubMedPMCPDF
- 56. Liu J, Zhou X, Li Q, Zhou SM, Hu B, Hu GW, et al. Role of phosphorylated HDAC4 in stroke-induced angiogenesis. Biomed Res Int 2017;2017:2957538.ArticlePubMedPMCPDF
- 57. Li L, Yang XJ. Molecular and functional characterization of histone deacetylase 4 (HDAC4). Methods Mol Biol 2016;1436:31-45.ArticlePubMed
- 58. Tao H, Shi KH, Yang JJ, Huang C, Zhan HY, Li J. Histone deacetylases in cardiac fibrosis: current perspectives for therapy. Cell Signal 2014;26:521-7.ArticlePubMed
- 59. Paroni G, Fontanini A, Cernotta N, Foti C, Gupta MP, Yang XJ, et al. Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol Cell Biol 2007;27:6718-32.ArticlePubMedPMCPDF
- 60. Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol 2005;25:2273-87.ArticlePubMedPMCPDF
- 61. Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res 2001;29:3439-47.ArticlePubMedPMC
- 62. Bolger TA, Zhao X, Cohen TJ, Tsai CC, Yao TP. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem 2007;282:29186-92.ArticlePubMed
- 63. Ciccarelli M, Vastolo V, Albano L, Lecce M, Cabaro S, Liotti A, et al. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia 2016;59:176-86.ArticlePubMedPDF
- 64. Paroni G, Mizzau M, Henderson C, Del Sal G, Schneider C, Brancolini C. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol Biol Cell 2004;15:2804-18.ArticlePubMedPMC
- 65. Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol 2005;25:8456-64.ArticlePubMedPMCPDF
- 66. Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem 2006;281:4423-33.ArticlePubMed
- 67. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721-32.ArticlePubMedPDF
- 68. Shi YH, Fang WG. Hypoxia-inducible factor-1 in tumour angiogenesis. World J Gastroenterol 2004;10:1082-7.ArticlePubMedPMC
- 69. Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 2001;15:2675-86.ArticlePubMedPMC
- 70. Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett 2009;583:55-60.PubMed
- 71. Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell 2010;38:864-78.PubMed
- 72. Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res 2006;66:8814-21.PubMed
- 73. Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, et al. HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem 2011;286:38095-102.ArticlePubMedPMC
- 74. Chen S, Sang N. Hypoxia-inducible factor-1: a critical player in the survival strategy of stressed cells. J Cell Biochem 2016;117:267-78.ArticlePubMedPMC
- 75. Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev 2009;23:223-35.ArticlePubMedPMC
- 76. Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev 2006;20:473-85.ArticlePubMedPMC
- 77. Kaowinn S, Kaewpiboon C, Koh SS, Kramer OH, Chung YH. STAT1-HDAC4 signaling induces epithelial-mesenchymal transition and sphere formation of cancer cells overexpressing the oncogene, CUG2. Oncol Rep 2018;40:2619-27.ArticlePubMedPMC
- 78. Kaewpiboon C, Srisuttee R, Malilas W, Moon J, Oh S, Jeong HG, et al. Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol Med Rep 2015;11:2315-21.ArticlePubMed
- 79. Stronach EA, Alfraidi A, Rama N, Datler C, Studd JB, Agarwal R, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res 2011;71:4412-22.ArticlePubMedPMCPDF
- 80. Park EJ, Kim YM, Kim HJ, Chang KC. Degradation of histone deacetylase 4 via the TLR4/JAK/STAT1 signaling pathway promotes the acetylation of high mobility group box 1 (HMGB1) in lipopolysaccharide-activated macrophages. FEBS Open Bio 2018;8:1119-26.ArticlePubMedPMCPDF
- 81. Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, et al. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell 2009;35:806-17.PubMed
- 82. Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta 2012;1819:707-15.ArticlePubMed
- 83. Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008;16:1407-16.ArticlePubMedPMC
- 84. Mosley AL, Ozcan S. Glucose regulates insulin gene transcription by hyperacetylation of histone h4. J Biol Chem 2003;278:19660-6.ArticlePubMed
- 85. Cho HM, Seok YM, Lee HA, Song M, Kim I. Repression of transcriptional activity of forkhead box O1 by histone deacetylase inhibitors ameliorates hyperglycemia in type 2 diabetic rats. Int J Mol Sci 2018;19:3539.ArticlePubMedPMC
- 86. Zhao H, Shu L, Huang W, Song G, Ma H. Resveratrol affects hepatic gluconeogenesis via histone deacetylase 4. Diabetes Metab Syndr Obes 2019;12:401-11.PubMedPMC
- 87. Gong M, Yu Y, Liang L, Vuralli D, Froehler S, Kuehnen P, et al. HDAC4 mutations cause diabetes and induce β-cell FoxO1 nuclear exclusion. Mol Genet Genomic Med 2019;7:e602.ArticlePubMedPMCPDF
- 88. Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, et al. A hormone-dependent module regulating energy balance. Cell 2011;145:596-606.ArticlePubMedPMC
- 89. Wang B, Liu TY, Lai CH, Rao YH, Choi MC, Chi JT, et al. Glycolysis-dependent histone deacetylase 4 degradation regulates inflammatory cytokine production. Mol Biol Cell 2014;25:3300-7.ArticlePubMedPMC
- 90. Usui T, Okada M, Mizuno W, Oda M, Ide N, Morita T, et al. HDAC4 mediates development of hypertension via vascular inflammation in spontaneous hypertensive rats. Am J Physiol Heart Circ Physiol 2012;302:H1894-904.ArticlePubMed
- 91. Guo Y, Fang Q, Ma D, Yu K, Cheng B, Tang S, et al. Up-regulation of HO-1 promotes resistance of B-cell acute lymphocytic leukemia cells to HDAC4/5 inhibitor LMK-235 via the Smad7 pathway. Life Sci 2018;207:386-94.ArticlePubMed
- 92. Choi MC, Cohen TJ, Barrientos T, Wang B, Li M, Simmons BJ, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell 2012;47:122-32.ArticlePubMedPMC
- 93. Yang Q, Tang J, Pei R, Gao X, Guo J, Xu C, et al. Host HDAC4 regulates the antiviral response by inhibiting the phosphorylation of IRF3. J Mol Cell Biol 2019;11:158-69.ArticlePubMedPDF
- 94. Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol 2011;32:335-43.ArticlePubMed
- 95. O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors: redefining innate immunity. Nat Rev Immunol 2013;13:453-60.ArticlePubMedPDF
- 96. Rossol M, Heine H, Meusch U, Quandt D, Klein C, Sweet MJ, et al. LPS-induced cytokine production in human monocytes and macrophages. Crit Rev Immunol 2011;31:379-446.ArticlePubMed
- 97. Abu-Farha M, Tiss A, Abubaker J, Khadir A, Al-Ghimlas F, Al-Khairi I, et al. Proteomics analysis of human obesity reveals the epigenetic factor HDAC4 as a potential target for obesity. PLoS One 2013;8:e75342.ArticlePubMedPMC
- 98. Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med 2005;11:191-8.ArticlePubMedPDF
- 99. Yang Q, Tang J, Xu C, Zhao H, Zhou Y, Wang Y, et al. Histone deacetylase 4 inhibits NF-κB activation by facilitating IκBα sumoylation. J Mol Cell Biol 2020;12:933-45.ArticlePubMedPDF
- 100. Denys A, Udalova IA, Smith C, Williams LM, Ciesielski CJ, Campbell J, et al. Evidence for a dual mechanism for IL-10 suppression of TNF-alpha production that does not involve inhibition of p38 mitogen-activated protein kinase or NF-kappa B in primary human macrophages. J Immunol 2002;168:4837-45.PubMed
- 101. Ozcan L, Ghorpade DS, Zheng Z, de Souza JC, Chen K, Bessler M, et al. Hepatocyte DACH1 is increased in obesity via nuclear exclusion of HDAC4 and promotes hepatic insulin resistance. Cell Rep 2016;15:2214-25.ArticlePubMedPMC
- 102. Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, et al. Endocrine regulation of energy metabolism by the skeleton. Cell 2007;130:456-69.ArticlePubMedPMC
- 103. Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R, et al. Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes 2011;60:2861-71.ArticlePubMedPMCPDF
- 104. Henriksson E, Sall J, Gormand A, Wasserstrom S, Morrice NA, Fritzen AM, et al. SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. J Cell Sci 2015;128:472-86.PubMedPMC
- 105. Weems JC, Griesel BA, Olson AL. Class II histone deacetylases downregulate GLUT4 transcription in response to increased cAMP signaling in cultured adipocytes and fasting mice. Diabetes 2012;61:1404-14.ArticlePubMedPMCPDF
- 106. Niu Y, Wang T, Liu S, Yuan H, Li H, Fu L. Exercise-induced GLUT4 transcription via inactivation of HDAC4/5 in mouse skeletal muscle in an AMPKα2-dependent manner. Biochim Biophys Acta Mol Basis Dis 2017;1863:2372-81.ArticlePubMed
- 107. Karamboulas C, Swedani A, Ward C, Al-Madhoun AS, Wilton S, Boisvenue S, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci 2006;119(Pt 20):4305-14.ArticlePubMedPDF
- 108. Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997;276:1404-7.ArticlePubMedPMC
- 109. Zhang L, Wang H, Zhao Y, Wang J, Dubielecka PM, Zhuang S, et al. Myocyte-specific overexpressing HDAC4 promotes myocardial ischemia/reperfusion injury. Mol Med 2018;24:37.ArticlePubMedPMCPDF
- 110. Zou G, Zhong W, Wu F, Wang X, Liu L. Catalpol attenuates cardiomyocyte apoptosis in diabetic cardiomyopathy via Neat1/miR-140-5p/HDAC4 axis. Biochimie 2019;165:90-9.ArticlePubMed
REFERENCES
Figure & Data
References
Citations
Citations to this article as recorded by 


 KDA
KDA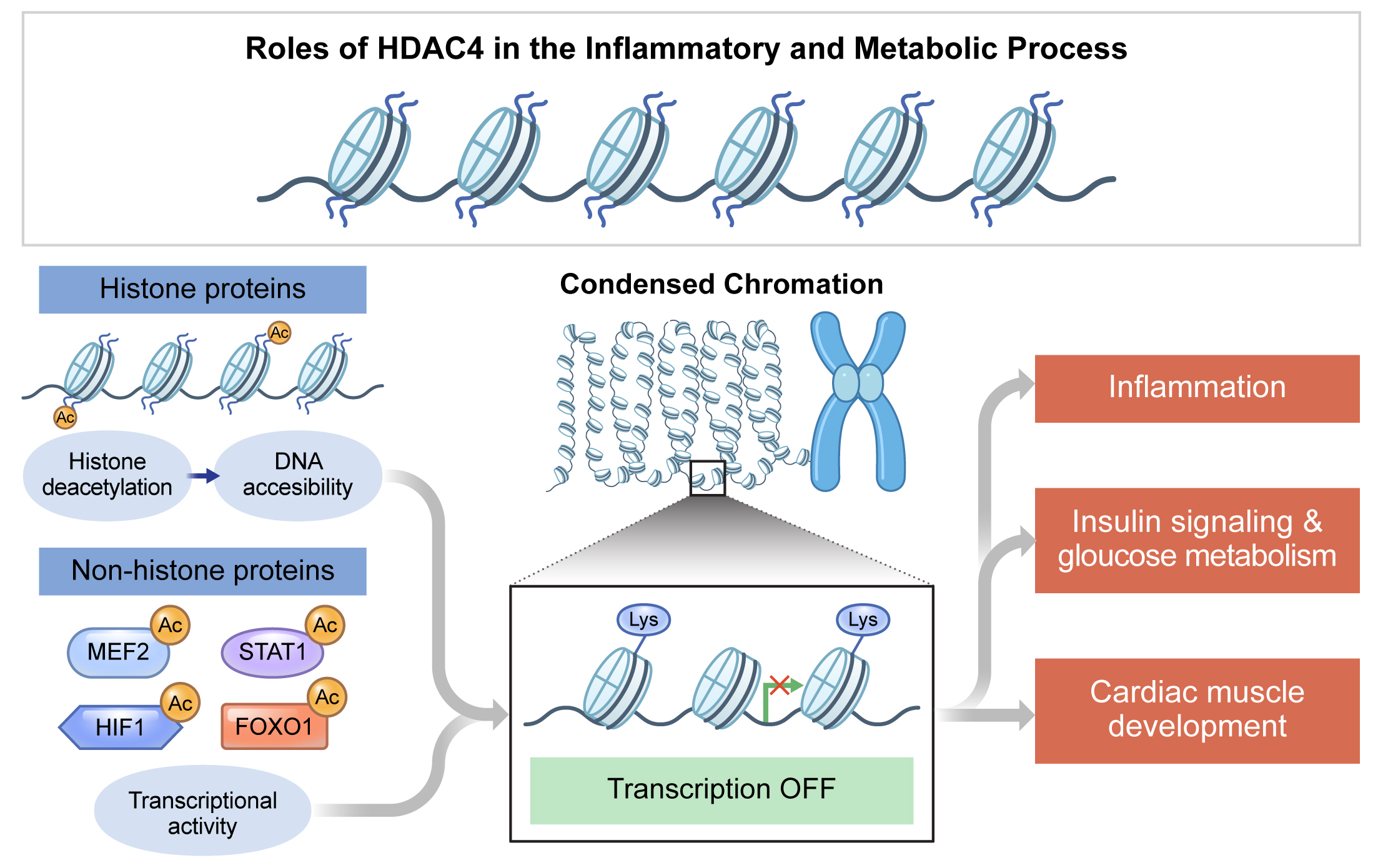
 PubReader
PubReader ePub Link
ePub Link Cite
Cite