MondoA Is Required for Normal Myogenesis and Regulation of the Skeletal Muscle Glycogen Content in Mice
Article information
Abstract
Background
Skeletal muscle is the largest tissue in the human body, and it plays a major role in exerting force and maintaining metabolism homeostasis. The role of muscle transcription factors in the regulation of metabolism is not fully understood. MondoA is a glucose-sensing transcription factor that is highly expressed in skeletal muscle. Previous studies suggest that MondoA can influence systemic metabolism homeostasis. However, the function of MondoA in the skeletal muscle remains unclear.
Methods
We generated muscle-specific MondoA knockout (MAKO) mice and analyzed the skeletal muscle morphology and glycogen content. Along with skeletal muscle from MAKO mice, C2C12 myocytes transfected with small interfering RNA against MondoA were also used to investigate the role and potential mechanism of MondoA in the development and glycogen metabolism of skeletal muscle.
Results
MAKO caused muscle fiber atrophy, reduced the proportion of type II fibers compared to type I fibers, and increased the muscle glycogen level. MondoA knockdown inhibited myoblast proliferation, migration, and differentiation by inhibiting the phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/Akt pathway. Further mechanistic experiments revealed that the increased muscle glycogen in MAKO mice was caused by thioredoxin-interacting protein (TXNIP) downregulation, which led to upregulation of glucose transporter 4 (GLUT4), potentially increasing glucose uptake.
Conclusion
MondoA appears to mediate mouse myofiber development, and MondoA decreases the muscle glycogen level. The findings indicate the potential function of MondoA in skeletal muscle, linking the glucose-related transcription factor to myogenesis and skeletal myofiber glycogen metabolism.
Highlights
• The glucose-related transcription factor, MondoA, was highly expressed in skeletal muscle, especially in type II muscle fibers.
• MondoA promotes mouse myofiber proliferation and differentiation, even affects morphology of skeletal muscle fiber.
• MondoA can decrease muscle glycogen level by regulating myocyte glucose uptake.
INTRODUCTION
As the largest tissue in the human body, skeletal muscle is a tissue with specific characteristics. Classically, skeletal muscle contracts and exerts physical force [12]. Additionally, skeletal muscle plays a key role in systemic energy metabolism, and this process can regulate the function of the skeletal muscle itself as well as the functions of other organs. Skeletal muscle energy metabolism, especially glucose metabolism, is critical for skeletal myogenesis, exercise endurance, and whole-body metabolism homeostasis.
MondoA, a glucose-sensing transcription factor, is predominantly highly expressed in skeletal muscle [34]. Unlike carbohydrate-response element-binding protein (ChREBP), another transcription factor in the Mondo family that is involved in glycolysis and lipogenesis [567], the target genes and functions of MondoA are incompletely understood. Several studies have demonstrated that thioredoxin-interacting protein (TXNIP) and arrestin domain-containing 4 (ARRDC4) are target genes of MondoA, making MondoA a negative regulator of cellular glucose uptake [89]. A recent study reported that MondoA directs myocyte fuel homeostasis by inhibiting glucose uptake and promoting lipogenesis [10]. Skeletal muscle is essential for regulating glucose metabolism as it is responsible for 70% of postprandial glucose uptake. However, the regulatory roles of MondoA in muscle fiber development and skeletal muscle glucose metabolism remain incompletely understood.
To determine the function of MondoA in skeletal muscle, we generated skeletal muscle-specific MondoA knockout (MAKO) mice and also knocked down MondoA using small interfering RNA (siRNA) in C2C12 muscle cells. MondoA was found to be highly expressed in type II (fast-twitch) glycolytic muscles and it was involved in myogenesis by regulating phosphoinositide 3-kinase (PI3K)/Akt signaling. The skeletal muscle of MAKO mice had increased glycogen levels due to increased glucose uptake.
METHODS
Animal experiments
C57BL/6J mice were purchased from Shanghai Slack in Laboratory Animal Ltd. (Shanghai, China). The mice were housed in a temperature- and humidity-controlled specific-pathogen-free environment with 12-hour light/dark cycles. They were fed normal chow with free access to water. Male mice at different ages (0.5 to 16 months old) were used for tissue collection.
To generate muscle-specific MAKO mice, a vector carrying two loxP sites flanking MondoA exon 2 was constructed using standard methods. The C57BL/6J embryonic stem (ES) cell line was used to generate gene-targeted ES cell clones, which were injected into C57BL/6J mice blastocysts to generate floxed mice. Floxed mice were crossed with transgenic mice expressing the Cre recombinase under the control of the creatine kinase MM isoenzyme promoter (CKMM-Cre). Polymerase chain reaction (PCR) was then performed to confirm the genotype of the muscle-specific MAKO mice. Wild-type (WT) littermate with same sex were used as control. All animal experiments were approved by the Shanghai Jiao Tong University School of Medicine Institutional Animal Care and Use Committee (approval number. XHEC-F-2020-003).
C2C12 cell line
Mouse C2C12 myoblasts (American Type Culture Collection, Manassas, VA, USA) were cultured in growth medium (GM) comprising Dulbecco's modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco, Waltham, MA, USA). When the C2C12 cell confluence was >80%, differentiation of the cells into myotubes was induced by replacing GM with differentiation medium (DM) comprising 2% horse serum in DMEM. DM was changed every 48 hours. The cells were cultured in a humidified incubator at 37℃ and 5% CO2.
In the in vitro experiments, C2C12 cells transfected with siRNA against MondoA (siMondoA) were compared to C2C12 cells transfected with negative control (NC) siRNA. Additionally, to explore the influence of phosphatase and tensin homolog (PTEN), we compared C2C12 cells transfected with siRNA against PTEN (siPTEN)+siMondoA with C2C12 cells transfected with NC+siMondoA.
Quantitative real-time PCR
Isolated of total RNA from different tissues or cells and reverse transcription are performed as described previously [11]. For quantitative real-time PCR (qRT-PCR), StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was applied. qPCR primers are shown in Table 1. 18s rRNA was used as the endogenous control to normalize mRNA expression and the relative expression levels of mRNAs were evaluated using the 2-ΔΔCt method.
Western blotting
The protein lysates of cells or tissue specimens were prepared as described previously [1112]. The proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE, 7.5% gel). The proteins were immobilized onto polyvinylidene fluoride membrane and immunoblotted with the specific antibodies. Antibodies against MondoA and TXNIP was purchase from Proteintech (Rosemont, IL, USA). Phosphorylated-Akt (p-Akt; Thr308), p-Akt (Ser473), Akt, and β-actin were purchased from Cell Signaling Technology (Boston, MA, USA), myogenin (MyoG), myosin heavy chain (MyHC), glucose transporter 1 (GLUT1), GLUT4, and α-tubulin antibodies were purchased from was purchased from Santa Cruz Biotechnology (Dallas, TX, USA).
Immunofluorescence analysis
Muscle samples were fixed in 4% paraformaldehyde, embedded in paraffin, cut cross-sectionally at 4 mm thickness. Paraffin-embedded gastrocnemius muscles were cut into 4-µm sections and placed on glass slides. The sections were rehydrated with phosphate-buffered saline (PBS), blocked using 5% goat serum, and incubated overnight with anti-dystrophin and anti-fast MyHC (type II-MyHC) antibodies (Abcam, Cambridge, MA, USA) at 4℃ and then with a fluorescent secondary antibody for 1 hour at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Yeasen, Shanghai, China) for 5 minutes. Additionally, to assess the fiber cross-sectional area (CSA), images were acquired using a scanning confocal microscope with Leica Application Suite (LAS) Advanced Fluorescence (AF) Lite (Leica Microsystems, Wetzlar, Germany). The CSA of type I and II diaphragm skeletal muscle myofibers were calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
To similarly use immunofluorescence to assess the MyHC protein expression and the myotube fusion index during the differentiation of C2C12 cells, the cells at 6-day postdifferentiation were fixed with 4% paraformaldehyde for 15 minutes and permeabilized with 0.5% Triton X-100. The immunofluorescent staining process was the same as that described above.
Forced swimming test
After fasted for 12 hours, mice were placed in a transparent plastic bucket filled with water at a temperature of 28℃ and depth of 25 cm. Mice swam with a weight equivalent to 5% of body weight tied to tail. When a mouse stop swimming and could not keep the nostril above the water surface, pull it out of the bucket and estimated as the swimming time.
siRNA transfection
siRNAs specific for mouse MondoA or NC were synthesized by GenePharma Co. Ltd. (Shanghai, China). The siRNA sequences were as follows, siMondoA forward, 5′-GCCGUUCGGUGCU-GCUCAATT-3′ and reverse, 5′-UUGAGCAGCACCGAACGGCTT-3′ and siPTEN forward, 5′-AAGAU-CUUGACCAAU-GGCUAATT-3′ and reverse, 5′-UUAGCCAUUGGUCAAGA-UCUU TT-3′. siRNAs were transfected into the C2C12 myoblasts using Lipofectamine RNAiMAX (Invitrogen, Madison, WI, USA) according to the manufacturer's instructions. Cells were then incubated for 48 hours and media were replaced with DM. Efficiencies of MondoA knockdowns were determined by qRT-PCR and Western blotting.
Cell migration assay
Transwell chambers (8 µm pore; Becton Dickinson, Franklin Lakes, NJ, USA) were placed into wells. The transfected myoblasts suspended in DMEM containing no FBS were seeded on the top chambers (5×104 cells/well), while DMEM with 10% FBS and was added to the bottom. After 16 hours incubation, the non-migrated myoblasts remained in the top layers were removed by cotton swab, and the migrated myoblasts in the bottom layers were fixed and stained with crystal violet to visualize the nuclei. Migrated myoblasts were photographed using an inverted optical microscope and counted by the ImageJ software.
Periodic acid-Schiff staining
For glycogen visualization, the sections were de-paraffinized, rehydrated, rinsed in deionized water, and then incubated in periodic acid for 7 minutes, Schiff's reagent for 15 minutes, hematoxylin for 1 minute, and acid alcohol differentiation solution for 3 seconds. Finally, dehydration, clearing, and mounting were performed.
Glycogen measurements
Muscle glycogen content was determined using a Glycogen Test Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Briefly, the frozen muscle samples were weighed and placed on ice. Next, 0.4 M KOH was added to the samples (3:1 v/w) and they were then boiled at 100℃ for 20 minutes. Distilled water (16:1 v/w) was added to create the glycogen test solution. For measurement, 100 µL glycogen test solution (or distilled water) and 900 µL distilled water were mixed together in a tube and 2 mL chromogenic agent dissolved in 95% to 98% H2SO4 was then added. The tube was then incubated at 100℃ for 5 minutes. After cooling, the absorbance was read on a spectrophotometer at 620 nm.
For analysis of the glycogen content of C2C12 cells, cells cultured in 10-cm dish after transfection with siMondoA or NC at 6-day postdifferentiation were washed with PBS and then treated with 225 µL of 0.4 M KOH at 100℃ for 20 minutes. Glycogen test solution was created by adding 225 µL distilled water. Glycogen measurement was then performed as for the muscle samples.
Glucose uptake
Glucose uptake by C2C12 myoblasts was assessed using a Glucose Test Kit. C2C12 myoblasts were transfected with siMondoA or NC for 48 hours and differentiation was induced using DM. After differentiation for 6 days, the culture medium was collected. Next, 10 µL culture medium and 1,000 µL test solution were mixed and cultured at 37℃ for 15 minutes. The absorbance was read on a spectrophotometer at 505 nm. The difference in absorbance between culture medium from siRNA-transfected and NC-transfected cells indicated the total glucose uptake of the cells per well. The corresponding mean glucose uptake per cell was defined as the ratio of the total glucose uptake to the number of cells per well.
Statistical analysis
All experiments were performed in triplicate. Data were analyzed using GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA, USA) and are expressed as mean±standard error of the mean. Statistical differences were assessed by Student's t-test. P<0.05 was considered to indicate a significant difference.
RESULTS
Tissue expression profile of MondoA and its expression pattern during C2C12 proliferation and differentiation
We first analyzed the expression profiles of 10 tissues collected from 8-month-old C57BL/6 mice. MondoA was highly expressed in skeletal muscle (Fig. 1A). We then analyzed MondoA expression in different muscle types. MondoA expression was significantly higher in the gastrocnemius, quadriceps, and extensor digitorum longus than the soleus muscle (which contains mainly type I muscle fibers, unlike the other muscles) (Fig. 1B, Supplementary Fig. 1). Thus, MondoA expression was higher in type II muscles than type I muscles. Regarding the 0.5 to 16-month-old mice, there were no differences in MondoA expression between juvenile and young adult mice, but there were reductions in the middle-aged 14 and 16-month-old mice (Supplementary Fig. 2).
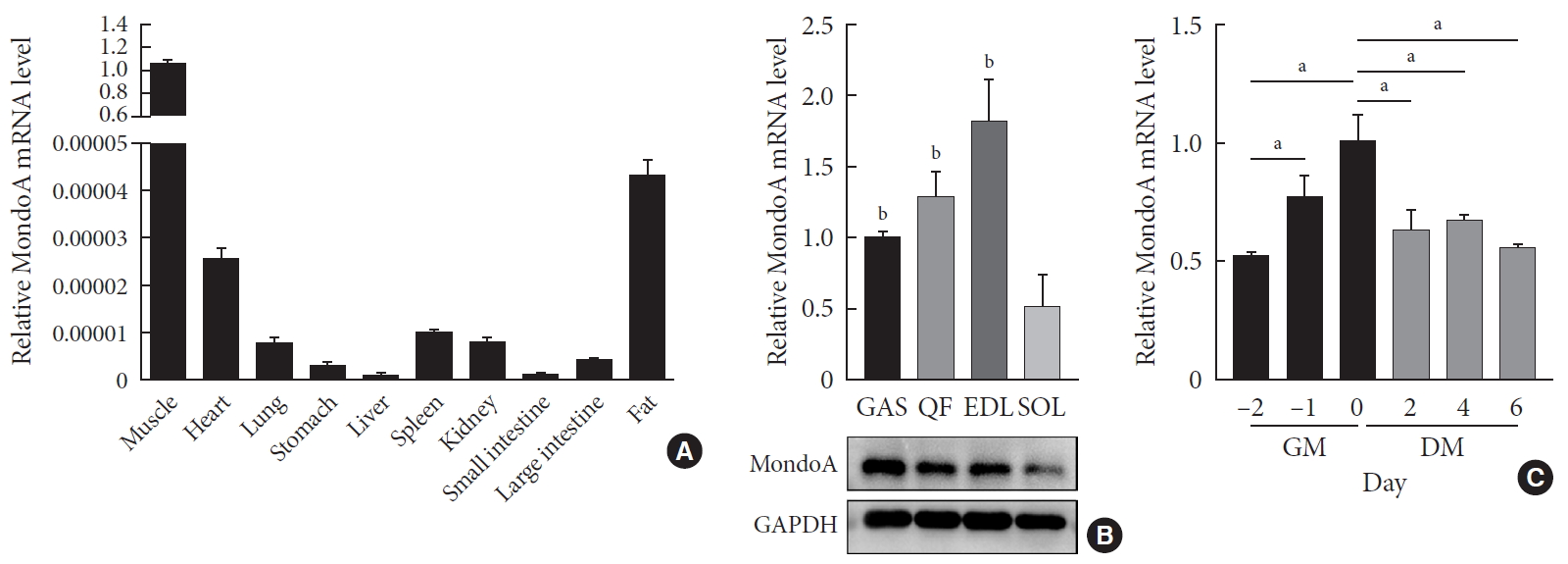
MondoA expression in various tissues and C2C12 cells. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of MondoA expression in various tissues (n=5 mice per group). (B) qRT-PCR (top) and Western blotting (bottom) analyses of MondoA expression in various muscles. (C) qRT-PCR analysis of MondoA expression in C2C12 cells cultured in growth medium (GM) and differentiation medium (DM) (experiments were performed in triplicate). Data represent mean±standard error of the mean. GAS, gastrocnemius; QF, quadriceps femoris; EDL, extensor digitorum longus; SOL, soleus; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. aP<0.05, bP<0.001.
Furthermore, to detect its role in muscle development, we assessed MondoA expression during C2C12 cell proliferation and differentiation. MondoA expression gradually increased during proliferation but exhibited no obvious change during differentiation, although the expression was lower in myotubes (after culture in DM) than in myoblasts (only cultured in GM) (Fig. 1C). This suggested that MondoA might play a significant role in skeletal muscle formation and function.
MAKO impairs morphology of skeletal muscle fiber
To assess whether MondoA contributes to the morphological characteristics of skeletal muscles, we generated mice with conditional MAKO in skeletal muscle. The gastrocnemius and quadriceps exhibited no differences in cross-section, weight, or color between MAKO and WT mice (Fig. 2A and B) and the swimming capacity in the two groups was comparable (Supplementary Fig. 3).
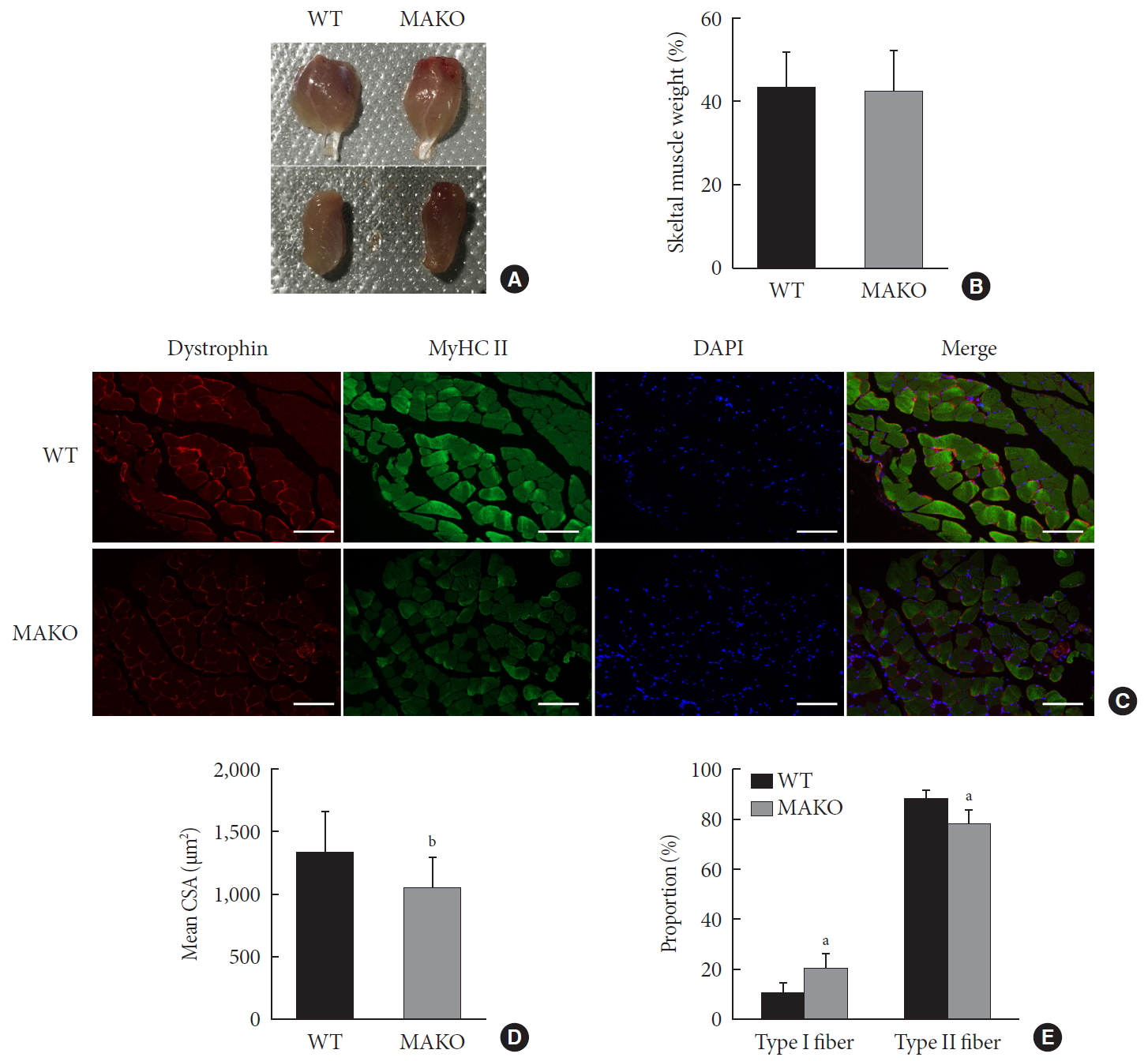
Muscle-specific MondoA knockout (MAKO) impairs myofiber size. (A) Morphology of gastrocnemius muscle (top) and quadriceps (bottom) from wild-type (WT) and MAKO mice. (B) Relative gastrocnemius muscle weight of WT and MAKO mice. (C) Representative fluorescent staining of myosin heavy chain (MyHC) type II fibers (green) and dystrophin (red) in gastrocnemius muscles. MyHC type I fibers are unstained (black) (n=5). Scale bar=100 µm. (D) Mean cross-sectional area (CSA; µm2) of type I and II diaphragm skeletal muscle myofibers. (E) Proportions (%) of muscle fiber types in entire cross-section of gastrocnemius muscle. CSA, cross-sectional area; DAPI, 4′,6-diamidino-2-phenylindole. aP<0.05, bP<0.001.
Additionally, the CSA of type I and II diaphragm myofibers from WT and MAKO mice was determined. The CSA in MAKO mice was reduced by 21% (Fig. 2C and D). The proportion of type II fibers was reduced by 9.7% in the MAKO gastrocnemius muscle, while the proportion of type I fibers was increased by 9.7% (Fig. 2E). The reduced mass in the MAKO muscles indicates that MAKO impairs the development of muscle fibers.
MAKO and MondoA knockdown inhibit myocyte proliferation and differentiation
As muscle fibers were thinner in MAKO mice, we speculated that MondoA may be involved in muscle cell proliferation or differentiation. We first investigated whether MondoA plays a role in myoblast proliferation. C2C12 cells were transfected with siMondoA or NC, and fewer colonies were formed after siMondoA transfection (Fig. 3A). The microscope images of the C2C12 myoblasts cultured in GM showed that siMondoA-transfected cells reached only 50% to 60% confluence, while the NC cells reached almost 100% confluence (Fig. 3B). Consistently, the numbers of siMondoA-transfected cells were significantly decreased compared to NC cells after 24, 48, and 72 hours, and the differences between the two groups grew larger over time (Fig. 3C). Additionally, the mRNA expression of cyclinD1 and E2, two cell proliferation activators [1314], also decreased after MondoA knockdown (Fig. 3D). Together, these results suggest that MondoA is essential for normal proliferation of C2C12 myoblasts.
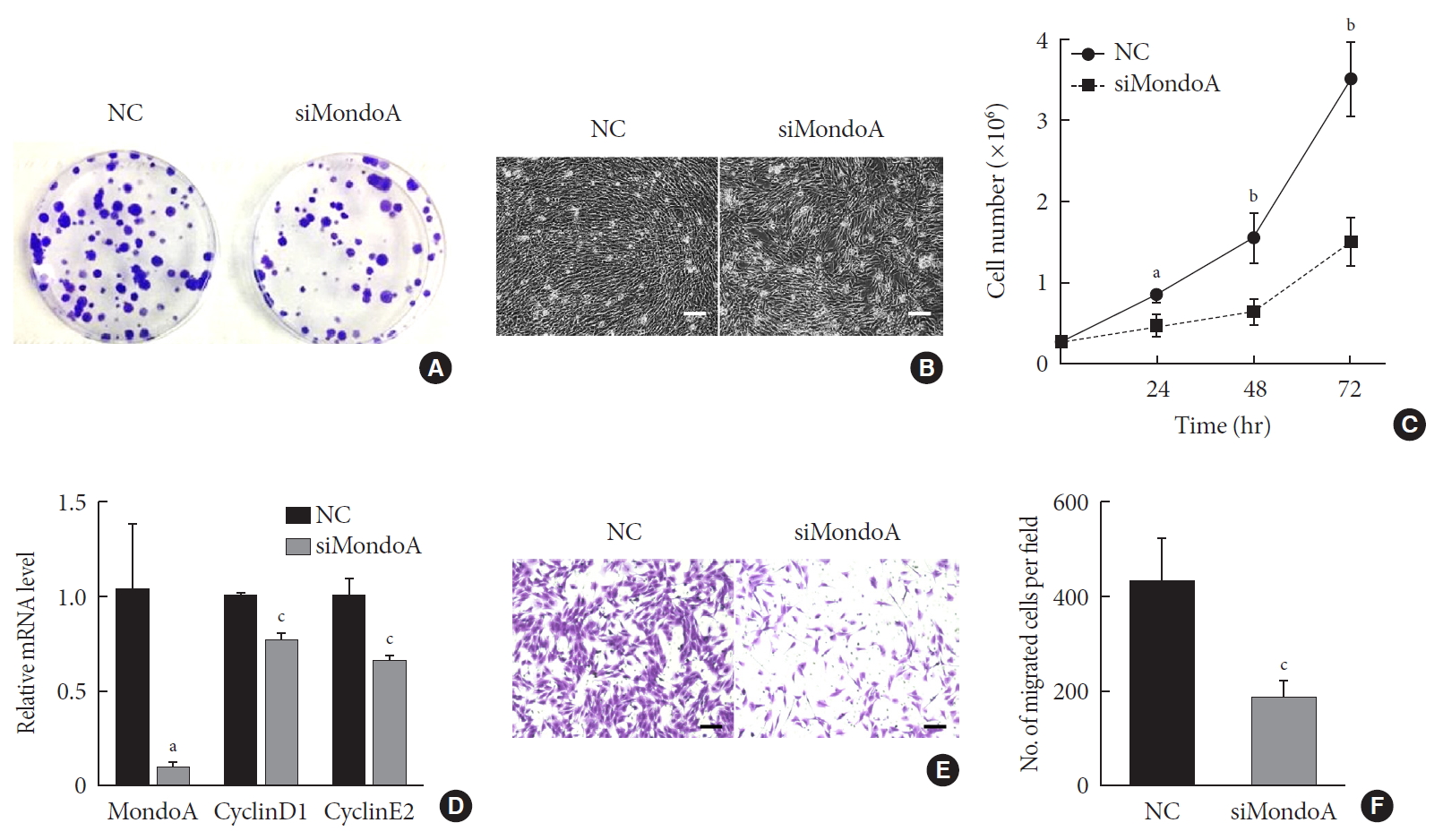
MondoA knockdown decreases C2C12 cell proliferation and migration. (A) Proliferation was analyzed by colony formation assay after 1.0×103 C2C12 myoblasts transfected with small interfering RNA against MondoA (siMondoA) or negative control (NC) were seeded and cultured for 7 days. (B) Proliferation was analyzed by microphotograph after transfection with siMondoA or NC for 72 hours. (C) Cell number was assessed after culture in growth medium for 12, 24, 36, and 72 hours after seeding 2.8×105 transfected C2C12 myoblasts. (D) Quantitative real-time polymerase chain reaction analysis of cyclinD1 and E2 in transfected C2C12 myoblasts. (E) Transwell assay of transfected C2C12 myoblasts (5×104 cells were seeded in the top chamber). Scale bar=100 µm. (F) Histogram showing the numbers of migrated myoblasts. aP<0.05, bP<0.01, cP<0.001.
Cell migration is an important process for cell development and maintenance of multicellular organogenesis, so we also examined the effect of MondoA knockdown on C2C12 migration. Transwell assays showed that MondoA knockdown in myoblasts decreased the migrated cell number by 50% compared with in the NC group (Fig. 3E and F). Hence, MondoA may promote C2C12 myoblast migration.
We then explored the effect of MondoA on myogenesis by inducing C2C12 myoblast differentiation. Microscope images showed less myotube formation in siMondoA-transfected cells than in NC cells (Fig. 4A). The mRNA expression of myogenic markers such as MyHC, MyoG, and insulin growth factor 2 (IGF2) increased with the differentiation induction time (Supplementary Fig. 4). C2C12 cell differentiation status at 6 days after inducing differentiation was then analyzed by immunofluorescence staining of MyHC. MondoA knockdown significantly decreased the MyHC protein expression, myogenic differentiation, and myotube fusion index (Fig. 4B and C). Additionally, MyHC, MyoG, and IGF2 mRNA levels at 6 days after differentiation were significantly decreased by 70% to 90% after MondoA knockdown (Fig. 4D). Consistently, protein levels of MyHC and MyoG were also decreased after 2 to 6 days of differentiation induction, especially after 6 days (Fig. 4E, Supplementary Fig. 5A). These results suggest that MondoA may facilitate C2C12 cell differentiation.
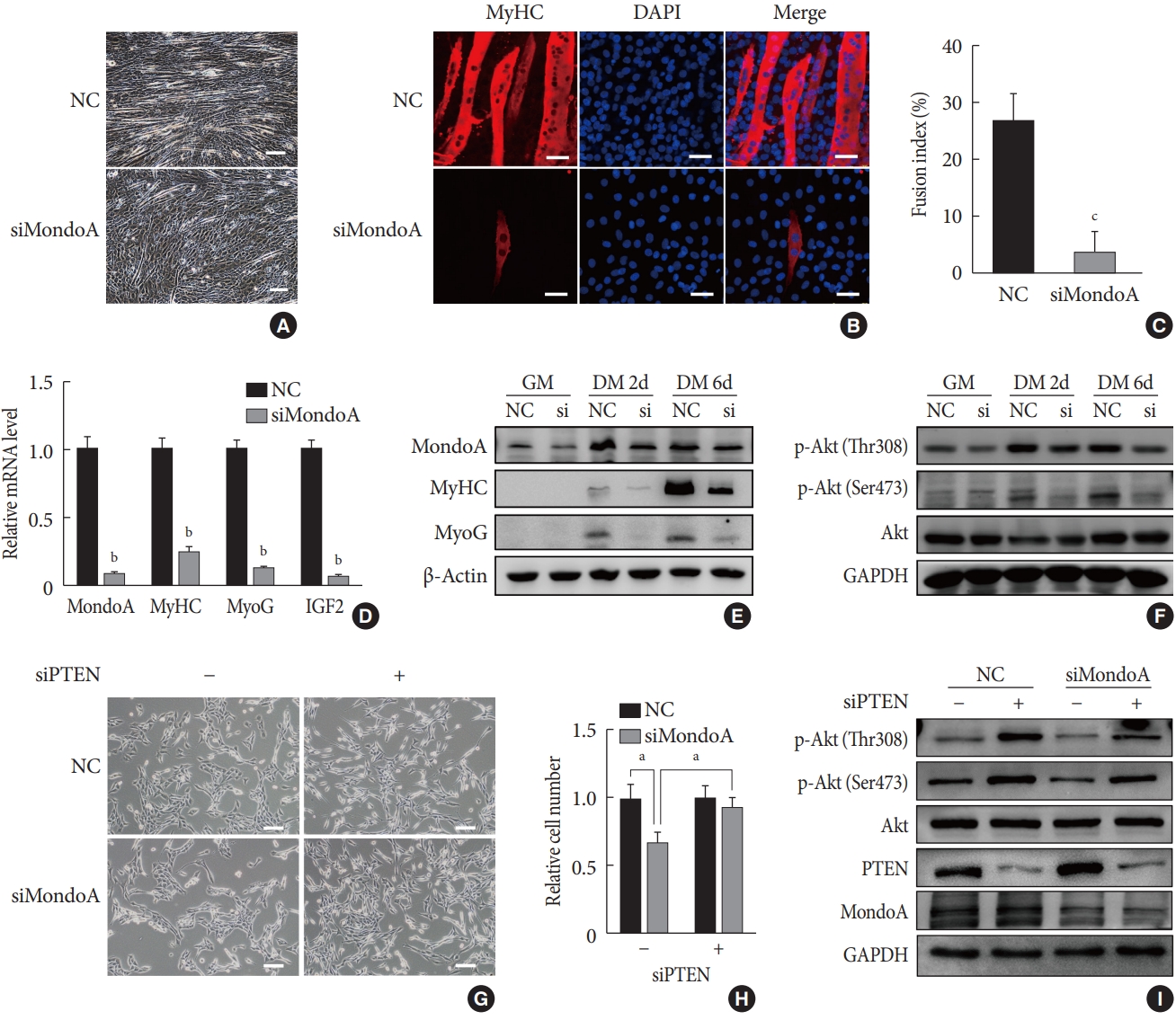
MondoA knockdown inhibited the myogenic differentiation of C2C12 myoblasts. (A) Microphotograph of C2C12 myoblasts, transfected with small interfering RNA (siRNA) against MondoA (siMondoA) or negative control (NC), at 6-day postdifferentiation. Scale bar=100 µm. (B) Immunostaining for myosin heavy chain (MyHC; red) and 4′,6-diamidino-2-phenylindole (DAPI) straining (blue) in C2C12 cells transfected with siMondoA or NC at 6-day postdifferentiation. The cells climbed to the carry sheet glass, were fixed with 4% paraformaldehyde, and were then stained with MyHC antibody (1:200). Scale bar=50 µm. (C) Fusion index of myotubes based on the ratio of the number of nuclei in MyHC-positive cells to the total number of nuclei (DAPI, blue), in at least five random microscopic fields. (D) Quantitative real-time polymerase chain reaction analysis of MondoA, MyHC, myogenin (MyoG), and insulin growth factor 2 (IGF2) at 6-day postdifferentiation in C2C12 cells transfected with siMondoA or NC. (E) Western blotting analysis of MondoA, MyHC, and MyoG at 0, 2, and 6-day postdifferentiation in C2C12 cells transfected with siMondoA or NC. (F) Western blotting analysis of key molecules in the phosphoinositide 3-kinase (PI3K)/Akt pathway at 0, 2, and 6-day postdifferentiation in C2C12 cells transfected with siMondoA or NC. (G) Proliferation was analyzed by microphotograph after transfection of cells with siMondoA and siRNA against phosphatase and tensin homolog (siPTEN) for 48 hours. −: siRNA against control. +: siPTEN. (H) Histogram showing the numbers of cells transfected with siMondoA and siPTEN for 48 hours. (I) Western blotting analysis of key molecules in the PTEN/PI3K/Akt pathway in C2C12 myoblasts transfected with siMondoA and siPTEN. GM, growth medium; DM, differentiation medium; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. aP<0.05, bP<0.01, cP<0.001.
"Updated on 30 September 2021"
MondoA knockdown inhibits myocyte proliferation and differentiation by blocking Akt activity
The PI3K/Akt pathway promotes myoblast proliferation and differentiation [1516]. We found that after 2 days of proliferation, 2 days of differentiation, and 6 days of differentiation, the protein level of p-Akt, especially p-Akt (Ser473) (the modification required for maximal Akt activity [17]), gradually increased after MondoA knockdown. Additionally, MondoA knockdown decreased the p-Akt (Ser473 and Thr308) levels during myoblast proliferation and differentiation (Fig. 4F, Supplementary Fig. 5B).
As MondoA induces phosphorylation of Akt (which is downstream in the PI3K/Akt pathway), we explored the mechanism. Previous research showed that PTEN is a negative regulator of the Akt pathway and skeletal muscle fiber development [18]. We found that siPTEN reversed the siMondoA-induced C2C12 cell proliferation inhibition (Fig. 4G and H). Western blotting indicated that there was an unexpected 2 to 3-fold increase in the p-Akt levels, especially the p-Akt (Ser473) levels, in siPTEN+siMondoA cells compared with NC+siMondoA cells (Fig. 4I, Supplementary Fig. 5C). Thus, MondoA knockdown inhibited proliferation and differentiation by promoting PTEN expression and thereby blocking Akt activity.
MAKO and MondoA knockdown increase muscle glycogen levels by regulating myocyte glucose uptake
We then used MAKO mice to determine whether MondoA contributes to skeletal muscle metabolism. Muscle glycogen metabolism is an important physiological function of skeletal muscle. Periodic acid-Schiff staining revealed that MAKO increased the glycogen content of skeletal muscle (Fig. 5A). Additionally, by quantifying the glycogen levels, we found that MAKO mice had a higher glycogen level compared with WT mice (Fig. 5B).
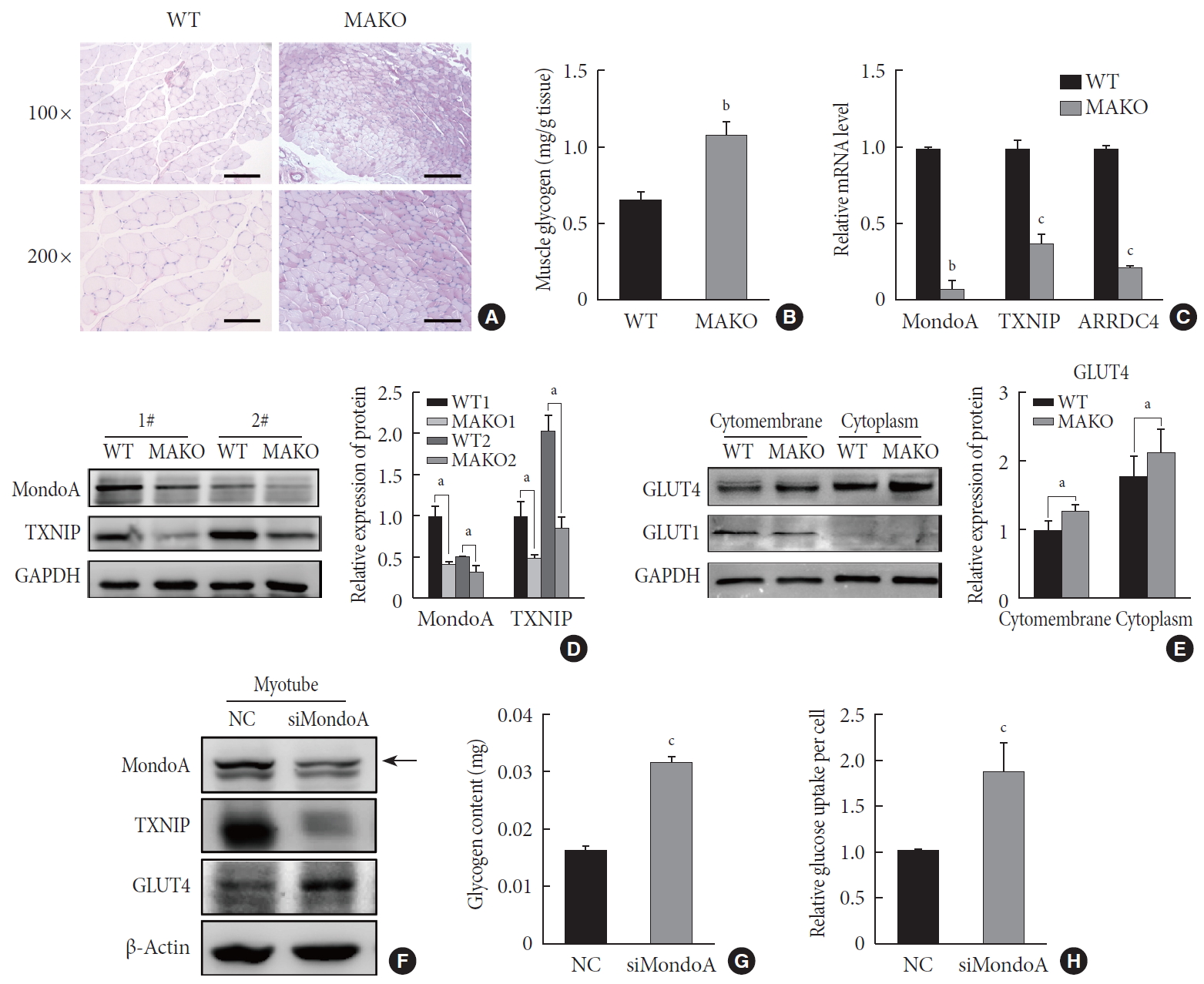
MondoA knockdown increases muscle glycogen level by promoting glucose uptake of skeletal muscle cells. (A) Periodic acid-Schiff (PAS) staining of gastrocnemius sections from wild-type (WT) and MondoA knockout (MAKO) mice. Scale bar=100 µm (top) or 200 µm (bottom). (B) Muscle glycogen content of gastrocnemius muscle (80 mg per group) from WT and MAKO mice (n=5). (C) Quantitative real-time polymerase chain reaction analysis of MondoA, thioredoxin-interacting protein (TXNIP), and arrestin domain-containing 4 (ARRDC4) in gastrocnemius muscle from WT and MAKO mice. (D) Western blotting analysis of MondoA and TXNIP in gastrocnemius muscle from WT and MAKO mice. (E) Western blotting analysis of glucose transporter 1 (GLUT1) and GLUT4 in the cell membrane and cytoplasm in the gastrocnemius muscle from WT and MAKO mice. (F) Western blotting analysis of GLUT1 and GLUT4 in C2C12 myotubes after transfection of C2C12 cells with small interfering RNA against MondoA (siMondoA) or negative control (NC). (G) Glycogen content of C2C12 myotubes cultured in 10-cm dishes after transfection with siMondoA or NC. (H) Relative glucose uptake of C2C12 myotubes after transfection with siMondoA or NC. aP<0.05, bP<0.01, cP<0.001.
The target genes of MondoA, TXNIP, and ARRDC4, have been considered as potent negative regulators of glucose uptake [192021]. We found that TXNIP and ARRDC4 mRNA and protein levels were reduced in the skeletal muscle of MAKO mice (Fig. 5C and D). Next, we determined the protein levels of GLUT1 and GLUT4, two essential transporters for glucose entry [22]. The protein levels of GLUT4 significantly increased in the skeletal muscle membrane and cytoplasm of MAKO mice compared to WT mice, while the GLUT1 level exhibited no difference (Fig. 5E, Supplementary Fig. 6A).
We hypothesized that MAKO improved the muscle glycogen level by regulating glucose uptake by myocytes. To test the hypothesis and determine whether the effects conferred by MAKO are relevant in vitro, we used siMondoA-transfected C2C12 cells. MondoA knockdown reduced TXNIP expression and increased GLUT4 expression in C2C12 myotubes (Fig. 5F, Supplementary Fig. 6B). Additionally, the glycogen content of myotubes significantly increased (Fig. 5G). These results were all consistent with the in vivo results. Next, we found that MondoA knockdown increased glucose uptake by about 1.87-fold in C2C12 myotubes (Fig. 5H). Taken together, these results demonstrate that MondoA may inhibit glycogen content by limiting glucose uptake by regulating TXNIP and GLUT4 expression.
DISCUSSION
In this study, we investigated the effect of MondoA on skeletal muscle, which is essential for glucose uptake and physical activity. MondoA was highly expressed in mouse skeletal muscle, but not in other metabolic tissues such as liver and adipose tissue. These results are consistent with previous research that showed that MondoA is enriched in skeletal muscle, unlike ChREBP, which is another Mondo family transcription factor [38]. Skeletal muscle is made up of fibers and it has distinctive metabolic and contractile properties. Skeletal muscle fibers are classified into two subtypes based on the expression of MyHC isoforms: type I (slow-twitch and oxidative) and type II (fast-twitch and glycolytic) [23]. We found that type II muscles exhibited higher MondoA expression than the type I muscle (soleus muscle), indicating that MondoA may be involved in glycolysis regulation in skeletal muscle. Further studies should assess the precise roles of MondoA in skeletal muscle biology.
The MAKO mice had atrophied muscle fibers with a decreased CSA, indicating that MAKO affected myofiber formation. We also found that MondoA was decreased in aging mice, in which muscle atrophy is one of the most distinctive features. These results indicated that MondoA is critical in mouse myofiber development. Previous studies showed similar the fiber-type distribution and ratio between type I and type II after MondoA global knockout [24]. Surprisingly, our study showed a reduced proportion of type II fibers compared to type I fibers after muscle-specific MondoA-knockdown. The difference provides evidences showing the essential role of MondoA in skeletal muscle. Further studies on its effect on muscle fiber size and subtype are essential.
Muscle development is essential for increasing muscle mass, determining the ratio of muscle fiber types, and certain muscle pathological changes [25]. To our knowledge, skeletal muscle development is an orderly process that can be divided into several steps including myoblast formation, proliferation, differentiation, and fusion into myotubes [26]. MondoA gradually increased during proliferation, and MondoA knockdown decreased C2C12 cell proliferation. Many studies have shown that cyclinD1 and E2 can influence cell proliferation by changing activity in the G0/G1 stage [2728]. Consistently, we also found that cyclinD1 and E2, which are involved in cell proliferation regulation, were downregulated after MondoA knockdown. Our data indicates that MondoA promotes myocyte proliferation via cell cycle regulation. Intercellular connection is needed for myogenesis induction [29]. MondoA-depletion induced weakened migration ability, the cell–cell contact of C2C12 cells, impacting C2C12 myoblasts proliferation and even terminal differentiation.
Few studies have explicitly reported that MondoA is involved in myoblast differentiation. The myogenic genes MyHC, MyoG, and IGF2 are necessary for driving early and late skeletal muscle differentiation [3031]. We demonstrated that the transcription factor MondoA promotes myoblast differentiation by increasing the expression of these myogenic genes. This explains how MondoA knockdown decreased the proliferation and differentiation of C2C12 cells. To a certain extent, the muscle atrophy in the MAKO mice may be attributable to the inhibited myoblast proliferation and differentiation.
PI3K/AKT signaling has been reported to be involved in myogenesis [1516]. Reduction of p-Akt has been shown to inhibit muscle cell proliferation [32]. Additionally, inhibition of Akt activation in myoblasts has been shown to effectively activate myogenic differentiation [33]. Similarly, we observed that p-Akt was decreased in myoblasts and myotubes following MondoA knockdown. Thus, our results suggested that MondoA may promote myoblast proliferation and differentiation by activating the Akt pathway. Previous studies have shown that PTEN acts as a negative regulator of the Akt pathway and skeletal muscle fiber development [18343536]. Yue et al. [3435] demonstrated that PTEN deletion alters muscle development. As shown in Fig. 4, PTEN knockdown was sufficient to reverse the proliferation inhibition caused by MondoA knockdown in C2C12 cells. These findings suggest that the PTEN/Akt pathway may underlie MondoA-dependent muscle development.
Glucose metabolites are important for the regulation of local and systemic energy by skeletal muscle [2237]. Uniquely, skeletal muscle can store excess glucose as glycogen after a meal, which can be rapidly decomposed to create glucose and generate energy [38]. Muscle glycogen is an essential factor in exercise and glucose homeostasis. After sensing the intracellular glucose state, MondoA promotes the transcription of certain target genes and then regulates energy homoeostasis [39]. Recent studies have clarified that MondoA drives lipid accumulation in skeletal muscle and promotes insulin resistance [1040]. However, whether the deletion of MondoA in skeletal muscle drives alternate glycogen metabolism has not been assessed. We found that muscle glycogen in vivo was higher after MAKO. GLUT4-mediated glucose uptake is essential for glycogen synthesis in skeletal muscle cells [41] and TXNIP, a target gene of MondoA, inhibits glucose uptake [919]. Our result showed decreased TXNIP and increased GLUT4 expression after MAKO. The results were consistent with the previous finding that TXNIP inhibits GLUT4 expression [42]. Thus, MAKO led to TXNIP downregulation followed by the elimination of the inhibition of GLUT4 expression, which may have increased glucose uptake, leading to increased muscle glycogen in the MAKO mice. We found that the glycogen level and glucose uptake increased after MondoA knockdown in vitro, which was in accordance with the in vivo data and verified our hypothesis. However, the muscle weights did not increase in the MAKO mice despite the decreased CSA, perhaps due to atrophied muscle fibers with elevated muscle glycogen levels. Moreover, the increased glucose uptake and glycogen levels may have compensated for the thinner myofibers, leading to the undiminished exercise capacity in the MAKO mice. These observations indicate that elevated glycogen in MAKO mice is caused by downregulation of TXNIP followed by upregulation of GLUT4-induced glucose uptake.
In summary, MondoA appears to mediate myofiber formation and development in mice. Additionally, MondoA expression may be critical for regulating the muscle glycogen content. The molecular mechanisms might involve upregulation of TXNIP by MondoA and the resultant downregulation of GLUT4-induced glucose uptake. These findings provide insights into the broad role of MondoA in skeletal muscle development and glucose metabolism. Targeting the molecular function of MondoA in skeletal muscle may provide novel clues for the treatment of metabolic diseases.
ACKNOWLEDGMENTS
We would like to thank Dr. Fan Hu, Sunyue He and Lei Liu for helping isolating and preserving skeletal muscle samples of mice in this research. This study was sponsored by the Shanghai Sailing Program (19YF1432200), National Natural Science Foundation of China (81370935, 81670743).
Notes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Conception or design: H.R., X.T., Q.S.
Acquisition, analysis, or interpretation of data: H.R., Y.L., Q.Z., Q.H., J.Z., K.W
Drafting the work or revising: H.R.
Final approval of the manuscript: H.R., K.W., Q.S.
References
SUPPLEMENTARY MATERIALS
Supplementary materials related to this article can be found online at https://doi.org/dmj.2019.0212.
Supplementary Fig. 1
Relative protein level of MondoA in various muscles. GAS, gastrocnemius; QF, quadriceps femoris; EDL, extensor digitorum longus; SOL, soleus. aP<0.05, bP<0.01, cP<0.001.
Supplementary Fig. 2
(A) Quantitative real-time polymerase chain reaction analysis, (B) Western blotting analysis, and (C) relative protein level of MondoA in skeletal muscle from mice of different ages. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. aP<0.05, bP<0.01, cP<0.001.
Supplementary Fig. 3
Exercise ability (assessed based on swimming time) of wild-type (WT) and MondoA knockout (MAKO) mice.
Supplementary Fig. 4
Expression levels of myogenic markers increased during differentiation. Relative mRNA levels of (A) myosin heavy chain (MyHC), (B) myogenin (MyoG), and (C) insulin growth factor 2 (IGF2) during culture in growth medium (GM) (−2, −1, and 0 day) and differentiation medium (DM) (2, 4, 6, and 8 days). aP<0.001.
Supplementary Fig. 5
(A) Relative protein levels of MondoA, myosin heavy chain (MyHC), and myogenin (MyoG) in C2C12 cells transfected with small interfering RNA against MondoA (siMondoA) or negative control (NC) at 0, 2, and 6-day postdifferentiation. (B) Relative protein levels of key molecules in the phosphoinositide 3-kinase (PI3K)/Akt pathway in C2C12 cells transfected with siMondoA or NC at 0, 2, and 6-day postdifferentiation. (C) Relative protein levels of key molecules in the phosphatase and tensin homolog (PTEN)/PI3K/Akt pathway in C2C12 myoblasts transfected with siMondoA. GM, growth medium; DM, differentiation medium; d, day. aP<0.05, bP<0.01, cP<0.001.
Supplementary Fig. 6
(A) Relative protein levels of glucose transporter 1 (GLUT1) in the cell membrane and cytoplasm in the gastrocnemius muscle from wild-type (WT) and MondoA knockout (MAKO) mice. (B) Relative protein levels of MondoA, thioredoxin-interacting protein (TXNIP), and GLUT4 in C2C12 myotubes after transfection of cells with small interfering RNA against MondoA (siMondoA) or negative control (NC). aP<0.01, bP<0.001.

