- Current
- Browse
- Collections
-
For contributors
- For Authors
- Instructions to authors
- Article processing charge
- e-submission
- For Reviewers
- Instructions for reviewers
- How to become a reviewer
- Best reviewers
- For Readers
- Readership
- Subscription
- Permission guidelines
- About
- Editorial policy
Articles
- Page Path
- HOME > Diabetes Metab J > Volume 34(5); 2010 > Article
-
Original ArticleThe Effect of Tribbles-Related Protein 3 on ER Stress-Suppressed Insulin Gene Expression in INS-1 Cells
- Young Yun Jang1, Nam Keong Kim1, Mi Kyung Kim1, Ho Young Lee1, Sang Jin Kim1, Hye Soon Kim1, Hye-Young Seo2, In Kyu Lee2, Keun Gyu Park1
-
Korean Diabetes Journal 2010;34(5):312-319.
DOI: https://doi.org/10.4093/kdj.2010.34.5.312
Published online: October 31, 2010
- 3,503 Views
- 32 Download
- 5 Crossref
1Department of Internal Medicine, Keimyung University School of Medicine, Daegu, Korea.
2Department of Internal Medicine, Kyungpook National University School of Medicine, Daegu, Korea.
- Corresponding author: Keun Gyu Park. Department of Internal Medicine, Keimyung University School of Medicine, 194 Dongsan-dong, Jung-gu, Daegu 700-712, Korea. kgpark@dsmc.or.kr
• Received: March 23, 2010 • Accepted: August 11, 2010
Copyright © 2010 Korean Diabetes Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
-
Background
- The highly developed endoplasmic reticulum (ER) structure in pancreatic beta cells is heavily involved in insulin biosynthesis. Thus, any perturbation in ER function inevitably impacts insulin biosynthesis. Recent studies showed that the expression of tribbles-related protein 3 (TRB3), a mammalian homolog of Drosophilia tribbles, in various cell types is induced by ER stress. Here, we examined whether ER stress induces TRB3 expression in INS-1 cells and found that TRB3 mediates ER stress-induced suppression of insulin gene expression.
-
Methods
- The effects of tunicamycin and thapsigargin on insulin and TRB3 expression in INS-1 cells were measured by Northern and Western blot analysis, respectively. The effects of adenovirus-mediated overexpression of TRB3 on insulin, PDX-1 and MafA gene expression in INS-1 cells were measured by Northern blot analysis. The effect of TRB3 on insulin promoter was measured by transient transfection study with constructs of human insulin promoter.
-
Results
- The treatment of INS-1 cells with tunicamycin and thapsigargin decreased insulin mRNA expression, but increased TRB3 protein expression. Adenovirus-mediated overexpression of TRB3 decreased insulin gene expression in a dose-dependent manner. A transient transfection study showed that TRB3 inhibited insulin promoter activity, suggesting that TRB3 inhibited insulin gene expression at transcriptional level. Adenovirus-mediated overexpression of TRB3 also decreased PDX-1 mRNA expression, but did not influence MafA mRNA expression.
-
Conclusions
- This study showed that ER stress induced TRB3 expression, but decreased both insulin and PDX-1 gene expression in INS-1 cells. Our data suggest that TRB3 plays an important role in ER stress-induced beta cell dysfunction.
- The endoplasmic reticulum (ER), a membrane component that is located near the nucleus, is an organelle where amino acids synthesized by mRNA become mature proteins after going through folding, assembly, glycosylation, disulfide bonding, and post-translational modifications [1]. ER stress is induced by a large influx of immature proteins that the ER cannot manage or the depletion of calcium by physiological or pathological environment [1-3]. The ER is well developed in endocrine cells such as pancreatic beta cells in which proteins are synthesize and secreted. Active insulin is made in the ER from preproinsulin, which is converted to proinsulin by post-translational modification and disulfide bond formation, and then removal of C-peptide [4,5]. Because the ER of pancreatic beta cells plays an important role in synthesizing and secreting active insulin, any ER stress by pathological conditions can impair the biosynthesis of insulin [6].
- When ER stress occurs, cellular defense mechanisms related to ER stress response are activated for the survival of the cells [1-3]. The ER stress response is made up of: 1) activation of protein kinase RNA (PKR)-like ER kinase (PERK) and reduction of mRNA to protein translation by phosphorylation of eIF2α (eukaryotic translation initiation factor 2α subunit) [7], 2) activation of inositol-requiring 1 (IRE1)/X-box binding protein 1 (XBP-1) and transcription factor (ATF6) through the increased expression of ER chaperones and subsequent increase of ER folding capacity [8,9], 3) ER stress-associated protein degradation, which degrades unfolded or improperly folded proteins [10], and 4) apoptosis by the activation of CCAAT/enhancer-binding homologous protein (CHOP) [2,11].
- In addition, many intracellular signal transduction systems are generated by ER stress; recent studies indicate that the expression of tribbles-related protein 3 (TRB3) is increased by ER stress-activated ATF4 and CHOP [12]. TRB3 contributes to insulin resistance by physically interrupting phosphorylation of Akt/PKB, which is important for insulin transduction [13]. During fasting, activation of cAMP response element binding protein (CREB) in liver induces the expression of peroxisome proliferator-activated (PPAR)-gamma coactivator-1α (PGC-1α) and caused gluconeogenesis. In this situation, activated CREB/PGC-1α increases TRB3 expression and causes insulin resistance by interrupting insulin action, which increases during food intake [14]. Additionally, TRB3 enters the nucleus by binding other transcription factors, which affects cellular growth, differentiation, and metabolism [13,15-17]. TRB3 in the nucleus is involved in the differentiation of adipocytes by activating CCAAT/enhancer-binding protein β (C/EBP) [18,19], and is also involved in lipid metabolism of adipocytes by activating E3 ligase constitutive photomorphogenic 1 (COP1) [20]. However, so far, there is little information on the role of TRB3 in pancreatic beta cells.
- The objectives of this study were to elucidate whether ER stress mediates the expression of TRB3 in INS-1 rat insulinoma cell line (INS-1 cell) and to determine the effect of TRB3 on the expression of insulin gene using adenovirus containing TRB3.
INTRODUCTION
- Materials
- Tunicamycin and thapsigargin used in the study were purchased from Sigma (St. Louis, MO, USA). The TRB3 antibodies used in the Western blot analysis were purchased from Calbiochem (La Jolla, CA, USA). Anti-rabbit antibodies and the radio-labeled probe ([α-32P]dCTP) was purchased from Amersham Bioscience (Little Chalfont, UK). Mouse TRB3 cDNA and recombinant TRB3 adenovirus was provided by Professor Seong-Hoi Koo of the Sungkyunkwan University College of Medicine.
- Cell incubation
- Insulin dependent rat host-cells, INS-1 cells, with subculture numbers between 20 and 30, were maintained in RPMI 1640 medium (Giboco-BRL, Grand Island, NY, USA) with 10% fetal bovine serum, 1 mM pyruvate, 10 mM HEPES, 50 mM 2-mercaptoethanol, 100 units/mL penicillin, 100 µg/mL streptomycin and incubated in 5% CO2/95% air at 37℃. The medium was changed every 3-4 days and was successively cultured using trypsin-EDTA.
- Preparation of recombinant adenovirus
- LacZ expressing adenovirus was used as a control for recombinant TRB3 adenovirus. The cDNA encoding a LacZ was inserted into the pAdTrack-CMV shuttle vector. The vector construct was then electroporated into BJ5138 cells, and a recombinant vector was generated using the AdEasy adenoviral vector system. The recombinant viruses were amplified in HEK-293 cells and purified by CsCl (Sigma) gradient centrifugation. Viral preparations were collected and desalted, and titers were determined using Adeno-X rapid titer (BD Bioscience, San Jose, CA, USA), according to the manufacturer's instructions.
- Northern blot analysis
- INS-1 cells were treated with tunicamycin and thapsigargin, and infected with adenoviral vectors expressing TRB3. Cells were harvested at the indicated times, total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and 20 µg of total RNA from each sample was used. The probes for insulin, PDX-1 and MafA were labeled with [α-32P]dCTP using a random-primer DNA-labeling system (Amersham Biosciences, Little Chalfont, UK).
- Western blot analysis
- INS-1 cells were treated with tunicamycin and thapsigargin for various times and then harvested in lysis buffer (50 mmol/L Tris-HCl [pH 8.0], 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Nonidet P-40, 0.25% Na-dexoycholate) containing proteinase inhibitors. The proteins were resolved by SDS-PAGE and then transferred electrophoretically to a polyvinyl difluoride membrane (Millipore, Bedford, MA, USA). The membrane was blocked by incubation in blocking buffer, incubated with anti-TRB3 antibody and was then developed using an ECL Western blot detection kit (Amersham Biosciences). The membrane was reblotted with anti-actin antibody to verify equal loading of protein in each lane. Densitometry was used to quantitate the results, using the digitalized scientific software program UN-SCAN-IT (Skik Scientific Co., Orem, UT, USA).
- Luciferase activity measurement
- INS-1 cells were plated at a density of 3 × 105 cell per well in a 12-well plate and subcultured for 2 days in INS-1 medium. The promoter constructs (300 ng/well) and other DNAs were transiently transfected by using Lipofectamine™ 2000 transfection reagent (Invitrogen). β-galactosidase plasmids were co-transfected as an internal control. Cells were transfected for 4 hours, washed to remove plasmids. Cells were harvested approximately 24 hours after transfection for luciferase and β-galactosidase assays. 20 µL of cell lysate containing 15 µg of protein was analyzed by using the Luciferase assay system according to the manufacturer's instructions (Promega, Madison, WI, USA). Luciferase activity was detected using a SIRUS Luminometer (Berthold, Pforzheim, Germany). The luciferase activity was normalized by using the β-galactosidase activity.
- Induction of ER stress
- Tunicamycin and thapsigargin were used to induce ER stress in INS cells. Insulin expression was measured after 24 hours of treatment with 2 µg/mL tunicamycin, and after 5 hours of treatment with 1 µM thapsigargin. Additionally, TRB3 protein expression was measured at an indicated time point after treatment of tunicamycin and thapsigargin.
- Statistical analysis
- The results are given as the average ± standard deviation, and Duncan's test was used to analyze variables. Cases with a P value lower than 0.05 were statistically significant for determination, and all trials were independently run over three times.
METHODS
- The effect of ER stress on insulin gene expression in INS-1 cells
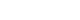
- To determine the effect of ER stress on the insulin gene expression, INS-1 cells were treated with the ER stress-inducing substances, tunicamycin and thapsigargin, and the changes in insulin mRNA were observed through Northern blot analysis. As shown in Fig. 1, control INS-1 cells showed high insulin mRNA expression, but INS-1 cells treated with tunicamycin and thapsigargin showed significantly decreased insulin mRNA expression.
- Effect of ER stress on TRB3 expression in INS-1 cells
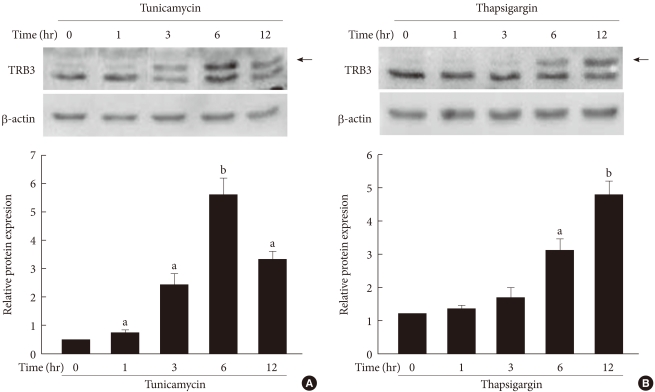
- To determine the effects of ER stress on the expression of TRB3 in INS-1 cells, cells were treated with tunicamycin and thapsargin and the changes in the TRB3 expression were measured after 1, 3, 6, and 12 hours using Western blot analysis. At baseline, the expression of TRB3 in INS-1 cells was weak. When treated with tunicamycin, TRB3 expression started to increase at 3 hours, reached its maximum rate at 6 hours, and continued to increase up to 12 hours. When treated with thapsigargin, TRB3 expression was slightly increased at 3 hours and continued to increase up to 12 hours (Fig. 2).
- Effects of TRB3 adenovirus on insulin gene expression
- To observe the effects of the ER-induced increase in TRB3 on the expression of the insulin gene in INS-1 cells, TRB3 overexpressing adenoviruses were prepared. After infecting INS-1 cells with TRB3-expressing adenovirus at varying concentrations, the proportional increase in TRB3 expression was confirmed (Fig. 3). Control INS-1 cells showed high expression of insulin mRNA. However, when TRB3 overexpression was induced with adenovirus, there was a dose-dependent decrease in insulin mRNA expression (Fig. 3). The decrease in insulin gene expression caused by TRB3 is not an effect of adenovirus because insulin mRNA expression did not decrease in response to the control LacZ-expressing adenovirus (Fig. 3).
- The effects of TRB3 on insulin promoter activity
- In order to determine the effect of TRB3 on insulin gene transcriptional activity, a human insulin promoter-expressing vector and TRB3 expressing vector were cotransfected in INS-1 cells. With this approach, we measured the activity of luciferase under the control of the insulin transcriptional promoter. As shown in Fig. 4, insulin promoter luciferase activity decreased in a dose-dependent manner with the TRB3 expressing vector. Thus, TRB3 appears to suppress gene expression that is regulated by the insulin gene transcription promoter region.
- The effects of TRB3 on the expression of MafA and PDX-1 in INS-1 cells
- Finally, we examined the effects of TRB3 on the expression of the pancreatic beta cell specific transcription factors, PDX-1 and MafA, which stimulate the expression of insulin mRNA. At baseline, INS-1 cells expressed high levels of PDX-1 and MafA mRNA, but when infected with adenovirus TRB3, the cells demonstrated significantly decreased PDX-1 mRNA expression but had no effect on the expression of MafA mRNA (Fig. 3).
RESULTS
- Several studies have demonstrated that in pancreatic beta cells with well developed ERs, beta cell dysfunction caused by ER stress can contribute to diabetes [21-25]. Initially, in obesity or type 2 diabetes, in order for the body to overcome insulin resistance in peripheral tissues and maintain normal blood glucose levels, hyperinsulinemia occurs. Thus, in long term continued cases of insulin resistance, demands for insulin synthesis and secretion continuously increase preproinsulin protein influx into the ER of pancreatic beta cells to maintain homoeostatic glucose metabolism and cause ER stress [2]. Therefore, the ER plays an important role in beta cell function. It can be assumed that beta cell dysfunction caused by ER stress can cause diabetes. Our previous study indicated that exposing INS-1 cells to high glucose conditions for a long time can reduce insulin gene expression due to glucose toxicity [26]. Additionally, chronic hyperglycemia causes ER stress in INS-1 cells and the pancreas islets of OLETF rats [6]. Our previous study indicated that the mechanism of ER stress-induced insulin gene suppression is related to the activation of ATF6 [6]. This study also showed that treating INS-1 cells with tunicamycin and thapsigargin results in a decrease in insulin gene expression, which agrees with results from our previous work. In addition, in this study, we confirmed that TRB3 reduces insulin gene expression in response to ER stress.
- TRB3 is expressed during fasting in the liver through the activation of the PPAR-α/PGC-1α pathway and causes insulin resistance by suppressing Akt/PKB activity [13,27]. Bi et al. [28] reported that TRB3 can cause insulin resistance in adipose tissues. In metabolic syndrome-induced rats, it has been reported that there is an increase in TRB3 and significant decrease in Akt phosphorylation in adipose tissue. Collectively, these data showed that TRB3 affects insulin resistance not only in liver tissue, but also in adipose tissue. It has been reported that ER stress induces the expression of TRB3 in a variety of cells. Corcoran et al. [29] reported that ER stress induces expression of TRB3 in the MCF-7 human breast cancer cell line, DU 145 human prostate cancer cell, and H1299 human lung cancer cell, etc. Additionally, it has been reported that tunicamycin increases the expression of TRB3 in human embryonic kidney cells and HepG2 liver cancer cells [12]. In this experiment, we confirmed that tunicamycin and thapsigargin induced ER stress and increased the expression of TRB3 in INS-1 rat insulinoma cells.
- Recently, Qian et al. [30] reported that the increased expression of TRB3 as a result of glucose toxicity and ER stress induces apoptosis and dysfunction of the pancreatic beta cells. In this study, we investigated the effect of ER stress-induced TRB3 expression on insulin gene expression. When TRB3 was overexpressed in INS-1 cells by infecting the cells with TRB3 expressing adenovirus, the expression of the insulin gene was significantly decreased. Additionally, it was confirmed that when human insulin gene transcription promoter and TRB3 are cotransfected in INS-1 cells, insulin gene transcription promoter activation is significantly decreased. These results suggest that TRB3 suppresses the expression of the insulin gene in at the transcriptional level. However, in order to confirm that ER stress-induced TRB3 expression is directly related to insulin gene suppression, it is necessary to elucidate whether suppression of TRB3 expression prevents ER stress-induced suppression of insulin gene expression.
- There are various transcription factors that regulate insulin gene expression at the insulin transcription promoter site. Among them, PDX-1 and MafA are important transcription factors that are involved in expression of insulin gene and are related to pancreatic beta cell function [31-36]. PDX-1 is an essential transcription factor for maintaining pancreatic development and beta cell differentiation and function [37]. MafA controls insulin transcription and also plays an important role in beta cell function [35]. Thus, inhibition of PDX-1 and MafA expression is closely related to beta cell dysfunction and diabetes [38]. In this experiment, we studied the effect that TRB3 on the expression of MafA and PDX-1. We confirmed that when TRB3 is overexpressed with adenovirus in INS-1 cells, MafA gene expression is not affected, while PDX-1 mRNA expression decreases. In order to verify whether the decrease in PDX-1 expression is directly related to the TRB3-dependent insulin gene suppression, additional studies are necessary to determine if the overexpression of PDX-1 offsets the effect of TRB3, which reduces insulin expression.
- In summary, factors causing ER stress in the INS-1 cells increase TRB3 expression. When TRB3 is overexpressed, insulin gene expression is suppressed. The TRB3-induced decrease in insulin gene expression is associated with the TRB3-induced suppression of PDX-1.
DISCUSSION
-
Acknowledgements
- This work was supported by Korean Diabetes Association Grant (2008) and the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2008-331-E00121).
ACKNOWLEDGMENT
- 1. Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol 2002;3:411-421. ArticlePubMedPDF
- 2. Oyadomari S, Araki E, Mori M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis 2002;7:335-345. PubMed
- 3. Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 2000;101:451-454. ArticlePubMed
- 4. Becker KL. Principles and practice of endocrinology and metabolism. 2001. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; p. 1296.
- 5. Dodson G, Steiner D. The role of assembly in insulin's biosynthesis. Curr Opin Struct Biol 1998;8:189-194. ArticlePubMed
- 6. Seo HY, Kim YD, Lee KM, Min AK, Kim MK, Kim HS, Won KC, Park JY, Lee KU, Choi HS, Park KG, Lee IK. Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases insulin gene expression via up-regulation of orphan nuclear receptor small heterodimer partner. Endocrinology 2008;149:3832-3841. ArticlePubMedPMCPDF
- 7. Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol 2002;18:575-599. ArticlePubMed
- 8. Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 1988;332:462-464. ArticlePubMedPDF
- 9. Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem 1998;273:33741-33749. ArticlePubMed
- 10. Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000;101:249-258. ArticlePubMed
- 11. Araki E, Oyadomari S, Mori M. Impact of endoplasmic reticulum stress pathway on pancreatic beta-cells and diabetes mellitus. Exp Biol Med (Maywood) 2003;228:1213-1217. ArticlePubMedPDF
- 12. Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 2005;24:1243-1255. ArticlePubMedPMC
- 13. Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 2003;300:1574-1577. ArticlePubMed
- 14. Okamoto H, Latres E, Liu R, Thabet K, Murphy A, Valenzeula D, Yancopoulos GD, Stitt TN, Glass DJ, Sleeman MW. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes 2007;56:1350-1356. ArticlePubMedPDF
- 15. Selim E, Frkanec JT, Cunard R. Fibrates upregulate TRB3 in lymphocytes independent of PPAR alpha by augmenting CCAAT/enhancer-binding protein beta (C/EBP beta) expression. Mol Immunol 2007;44:1218-1229. PubMed
- 16. Hegedus Z, Czibula A, Kiss-Toth E. Tribbles: novel regulators of cell function; evolutionary aspects. Cell Mol Life Sci 2006;63:1632-1641. ArticlePubMedPDF
- 17. Sung HY, Francis SE, Crossman DC, Kiss-Toth E. Regulation of expression and signalling modulator function of mammalian tribbles is cell-type specific. Immunol Lett 2006;104:171-177. ArticlePubMed
- 18. Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem 2007;282:24075-24082. PubMed
- 19. Bezy O, Vernochet C, Gesta S, Farmer SR, Kahn CR. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPbeta transcriptional activity. Mol Cell Biol 2007;27:6818-6831. PubMedPMCPDF
- 20. Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, Macleod IX, Liew CW, Kulkarni RN, Bain J, Newgard C, Nelson M, Evans RM, Yates J, Montminy M. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 2006;312:1763-1766. ArticlePubMed
- 21. Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099-1108. ArticlePubMed
- 22. Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, Urano F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab 2006;4:245-254. ArticlePubMed
- 23. Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell 2001;7:1153-1163. ArticlePubMed
- 24. Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med 2005;11:757-764. ArticlePubMedPDF
- 25. Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, MacAuley A, Goodman AG, LeBoeuf RC, Katze MG. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes 2005;54:1074-1081. PubMed
- 26. Park KG, Lee KM, Seo HY, Suh JH, Kim HS, Wang L, Won KC, Lee HW, Park JY, Lee KU, Kim JG, Kim BW, Choi HS, Lee IK. Glucotoxicity in the INS-1 rat insulinoma cell line is mediated by the orphan nuclear receptor small heterodimer partner. Diabetes 2007;56:431-437. ArticlePubMedPDF
- 27. Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med 2004;10:530-534. ArticlePubMedPDF
- 28. Bi XP, Tan HW, Xing SS, Wang ZH, Tang MX, Zhang Y, Zhang W. Overexpression of TRB3 gene in adipose tissue of rats with high fructose-induced metabolic syndrome. Endocr J 2008;55:747-752. ArticlePubMed
- 29. Corcoran CA, Luo X, He Q, Jiang C, Huang Y, Sheikh MS. Genotoxic and endoplasmic reticulum stresses differentially regulate TRB3 expression. Cancer Biol Ther 2005;4:1063-1067. ArticlePubMed
- 30. Qian B, Wang H, Men X, Zhang W, Cai H, Xu S, Xu Y, Ye L, Wollheim CB, Lou J. TRIB3 [corrected] is implicated in glucotoxicity-and endoplasmic reticulum-stress-induced [corrected] beta-cell apoptosis. J Endocrinol 2008;199:407-416. PubMed
- 31. Wang H, Maechler P, Ritz-Laser B, Hagenfeldt KA, Ishihara H, Philippe J, Wollheim CB. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J Biol Chem 2001;276:25279-25286. ArticlePubMed
- 32. Brissova M, Shiota M, Nicholson WE, Gannon M, Knobel SM, Piston DW, Wright CV, Powers AC. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J Biol Chem 2002;277:11225-11232. ArticlePubMed
- 33. Kulkarni RN, Jhala US, Winnay JN, Krajewski S, Montminy M, Kahn CR. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J Clin Invest 2004;114:828-836. ArticlePubMedPMC
- 34. Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes 2005;54:2586-2595. PubMed
- 35. Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol 2005;25:4969-4976. ArticlePubMedPMCPDF
- 36. Matsuoka TA, Kaneto H, Stein R, Miyatsuka T, Kawamori D, Henderson E, Kojima I, Matsuhisa M, Hori M, Yamasaki Y. MafA regulates expression of genes important to islet beta-cell function. Mol Endocrinol 2007;21:2764-2774. PubMed
- 37. Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994;371:606-609. ArticlePubMedPDF
- 38. Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev 1998;12:1763-1768. ArticlePubMedPMC
REFERENCES
Fig. 1The effects of tunicamycin and thapsigargin on insulin mRNA expression in INS-1 cells. Northern blot analysis of insulin mRNA expression in INS-1 cells treated with tunicamycin (A) and thapsigargin (B). INS-1 cells were treated with tunicamycin (2 µg/mL) for 24 hours or thapsigargin (1 µM) for 5 hours. 18S rRNA levels were analyzed as an internal control. Data in bar graph are the mean ± SEM of three independent measurements. aP < 0.01 and bP < 0.001 compared to control.
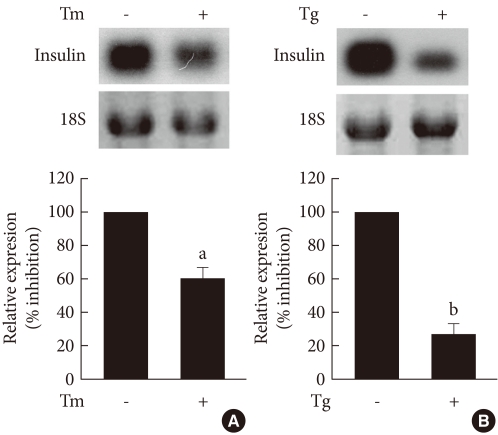

Fig. 2The effects of tunicamycin and thapsigargin on TRB3 protein expression in INS-1 cells. Western blot analysis of TRB3 protein expression in the presence of tunicamycin (A) and thapsigargin (B). INS-1 cells were incubated with tunicamycin (2 µg/mL) and thapsigargin (1 µM) for indicated times. β-actin protein levels were analyzed as an internal control. The data in bar graph are the mean ± SEM of three independent measurements. aP < 0.01 and bP < 0.001 compared to 0 hour.
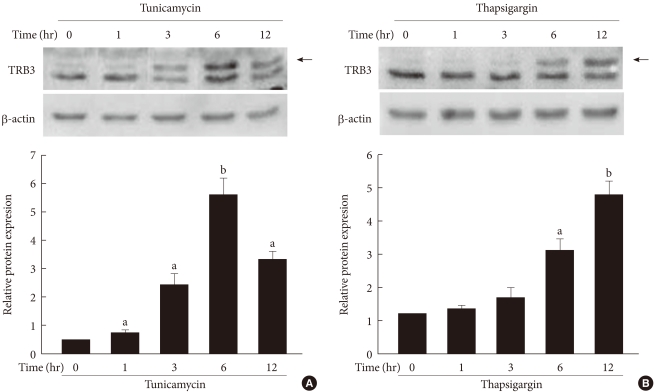

Fig. 3The effects of adenovirus-mediated overexpression of TRB3 on insulin, PDX-1 and MafA mRNA expression. Northern blot analysis of insulin, PDX-1 and MafA mRNA expression in INS-1 cells infected with adenovirus encoding TRB3 (Ad-TRB3). INS-1 cells were infected with the indicated doses of Ad-TRB3 or Ad-LacZ for 24 hours. 18S rRNA levels were analyzed as an internal control. Data in the bar graph are the mean ± SEM of three independent measurements. aP < 0.01 and bP < 0.001 compared to control.
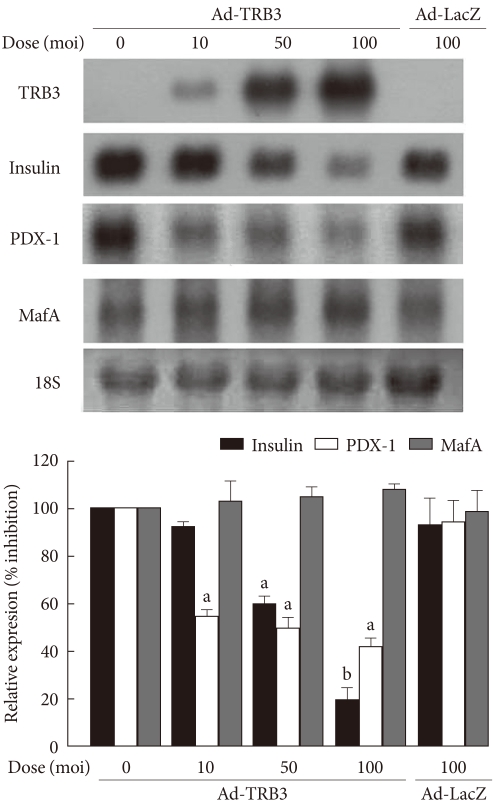

Fig. 4The effects of TRB3 on insulin promoter activity. INS-1 cells were cotransfected with the human insulin promoter (200 ng/well) and the indicated amounts of an expression vector for TRB3 for 24 hours. Data represent the mean ± SEM of three independent measurements. aP < 0.01 and bP < 0.001 compared to control.
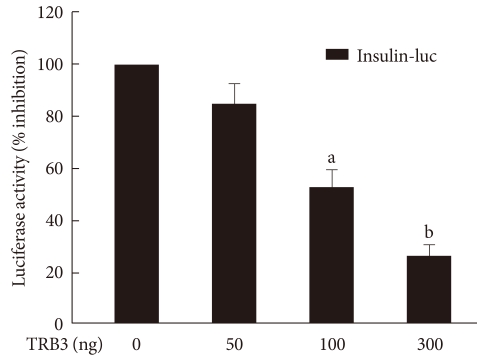

Figure & Data
References
Citations
Citations to this article as recorded by 

- Endoplasmic reticulum stress causes insulin resistance by inhibiting delivery of newly synthesized insulin receptors to the cell surface
Max Brown, Samantha Dainty, Natalie Strudwick, Adina D. Mihai, Jamie N. Watson, Robina Dendooven, Adrienne W. Paton, James C. Paton, Martin Schröder, James Arthur Olzmann
Molecular Biology of the Cell.2020; 31(23): 2597. CrossRef - PTB and TIAR binding to insulin mRNA 3′- and 5′UTRs; implications for insulin biosynthesis and messenger stability
Rikard G. Fred, Syrina Mehrabi, Christopher M. Adams, Nils Welsh
Heliyon.2016; 2(9): e00159. CrossRef - Asna1/TRC40 Controls β-Cell Function and Endoplasmic Reticulum Homeostasis by Ensuring Retrograde Transport
Stefan Norlin, Vishal S. Parekh, Peter Naredi, Helena Edlund
Diabetes.2016; 65(1): 110. CrossRef - Role of the Unfolded Protein Response inβCell Compensation and Failure during Diabetes
Nabil Rabhi, Elisabet Salas, Philippe Froguel, Jean-Sébastien Annicotte
Journal of Diabetes Research.2014; 2014: 1. CrossRef - Endoplasmic Reticulum Stress and Insulin Biosynthesis: A Review
Mi-Kyung Kim, Hye-Soon Kim, In-Kyu Lee, Keun-Gyu Park
Experimental Diabetes Research.2012; 2012: 1. CrossRef

 KDA
KDA PubReader
PubReader Cite
Cite